Detailed DSSR results for the G-quadruplex: PDB entry 1eeg
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 1eeg
- Class
- DNA
- Method
- NMR
- Summary
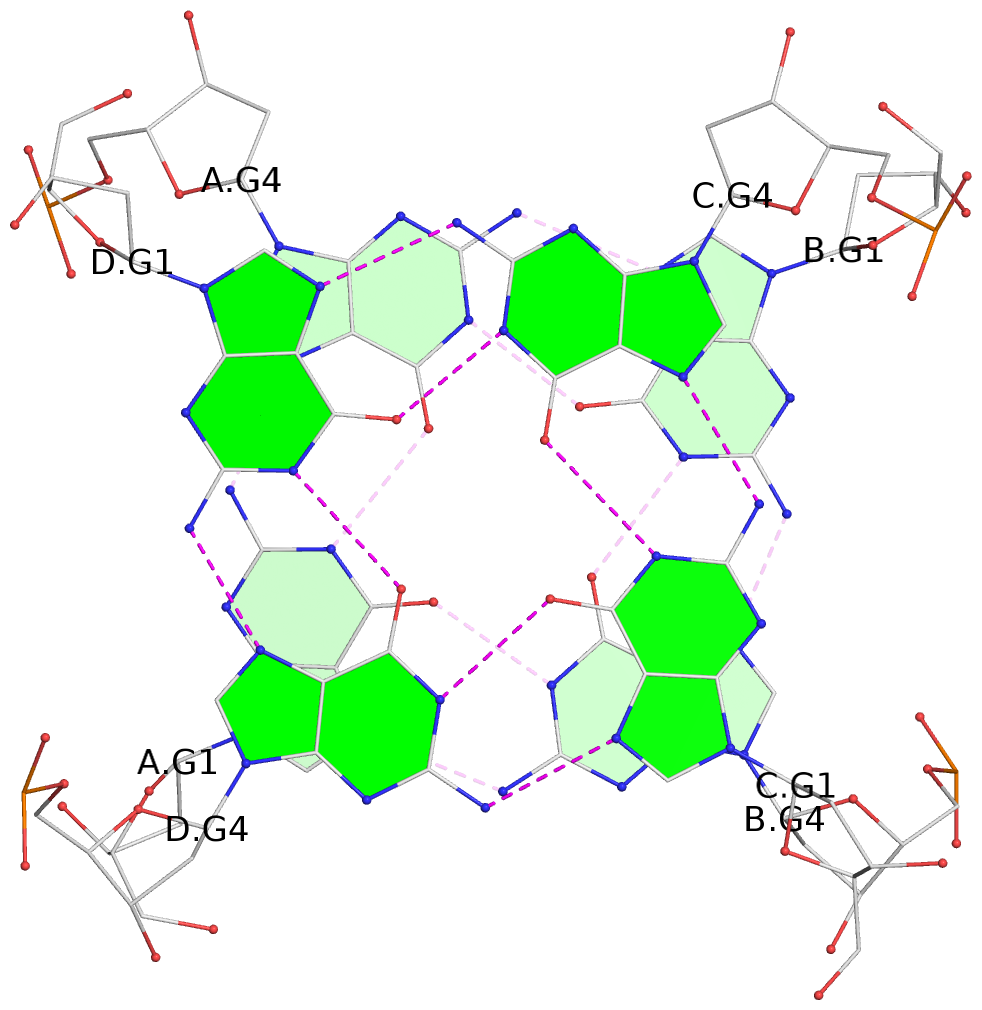
- A(gggg)a hexad pairing aligment for the d(g-g-a-g-g-a-g) sequence
- Reference
- Kettani A, Gorin A, Majumdar A, Hermann T, Skripkin E, Zhao H, Jones R, Patel DJ (2000): "A dimeric DNA interface stabilized by stacked A.(G.G.G.G).A hexads and coordinated monovalent cations." J.Mol.Biol., 297, 627-644. doi: 10.1006/jmbi.2000.3524.
- Abstract
- We report on the identification of an A.(G.G.G.G).A hexad pairing alignment which involves recognition of the exposed minor groove of opposing guanines within a G.G.G.G tetrad through sheared G.A mismatch formation. This unexpected hexad pairing alignment was identified for the d(G-G-A-G-G-A-G) sequence in 150 mM Na(+) (or K(+)) cation solution where four symmetry-related strands align into a novel dimeric motif. Each symmetric half of the dimeric "hexad" motif is composed of two strands and contains a stacked array of an A.(G.G.G.G).A hexad, a G.G.G.G tetrad, and an A.A mismatch. Each strand in the hexad motif contains two successive turns, that together define an S-shaped double chain reversal fold, which connects the two G-G steps aligned parallel to each other along adjacent edges of the quadruplex. Our studies also establish a novel structural transition for the d(G-G-A-G-G-A-N) sequence, N=T and G, from an "arrowhead" motif stabilized through cross-strand stacking and mismatch formation in 10 mM Na(+) solution (reported previously), to a dimeric hexad motif stabilized by extensive inter-subunit stacking of symmetry-related A.(G.G.G.G).A hexads in 150 mM Na(+) solution. Potential monovalent cation binding sites within the arrowhead and hexad motifs have been probed by a combination of Brownian dynamics and unconstrained molecular dynamics calculations. We could not identify stable monovalent cation-binding sites in the low salt arrowhead motif. By contrast, five electronegative pockets were identified in the moderate salt dimeric hexad motif. Three of these are involved in cation binding sites sandwiched between G.G.G. G tetrad planes and two others, are involved in water-mediated cation binding sites spanning the unoccupied grooves associated with the adjacent stacked A.(G.G.G.G).A hexads. Our demonstration of A.(G. G.G.G).A hexad formation opens opportunities for the design of adenine-rich G-quadruplex-interacting oligomers that could potentially target base edges of stacked G.G.G.G tetrads. Such an approach could complement current efforts to design groove-binding and intercalating ligands that target G-quadruplexes in attempts designed to block the activity of the enzyme telomerase.
- G4 notes
- 4 G-tetrads, 1 G4 helix, 2 G4 stems, 1 G4 coaxial stack, parallel(4+0), UUUU, coaxial interfaces: 5'/5'
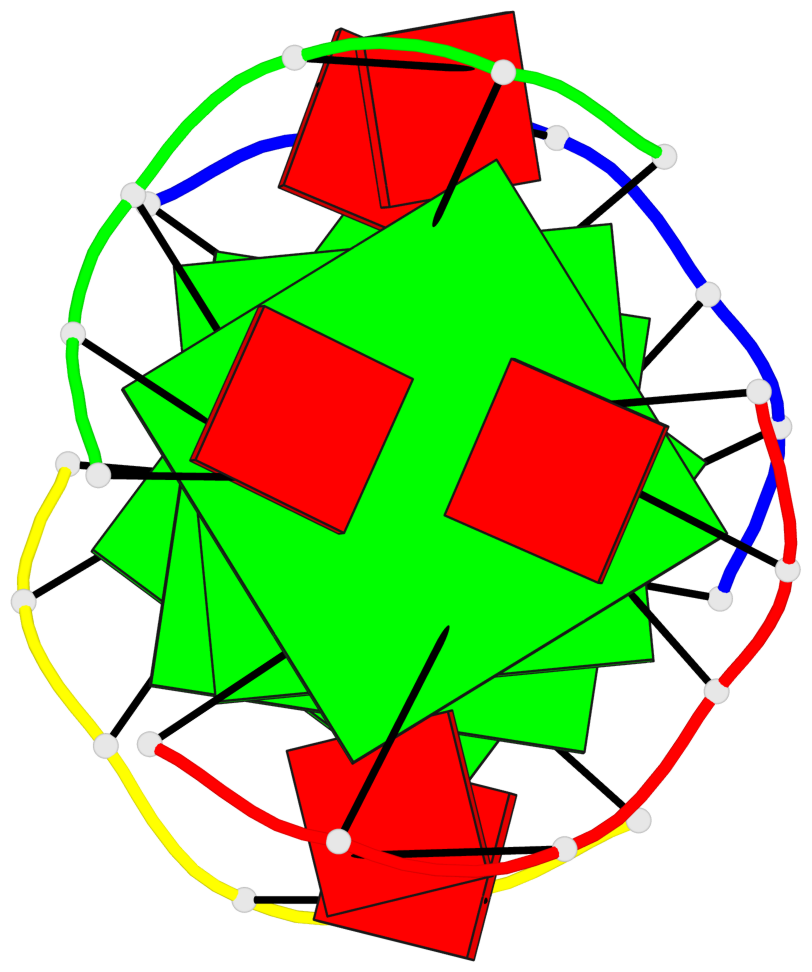
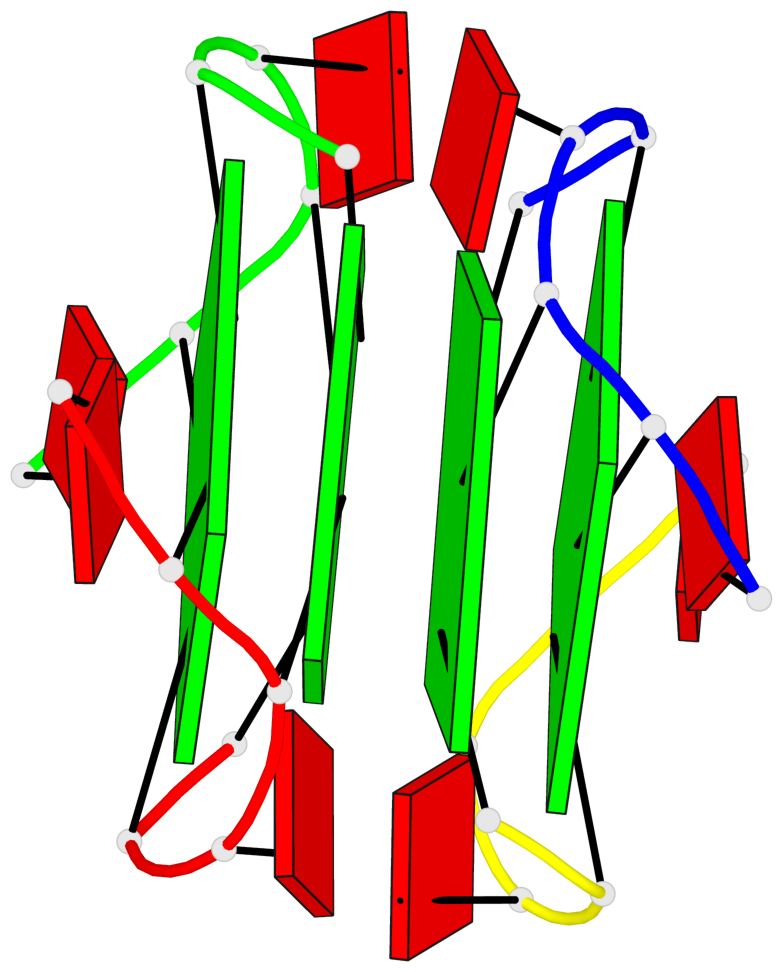
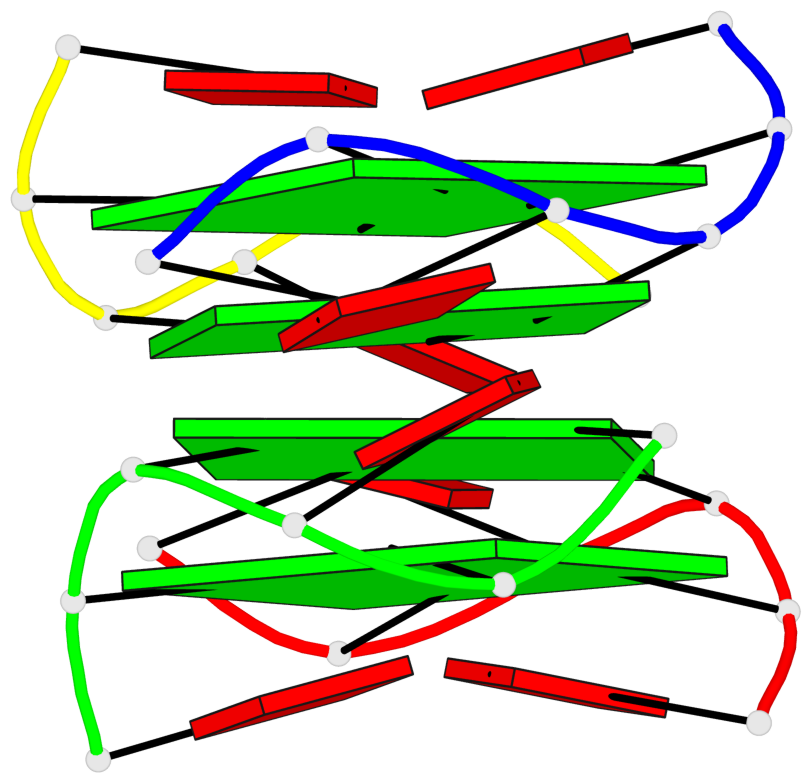
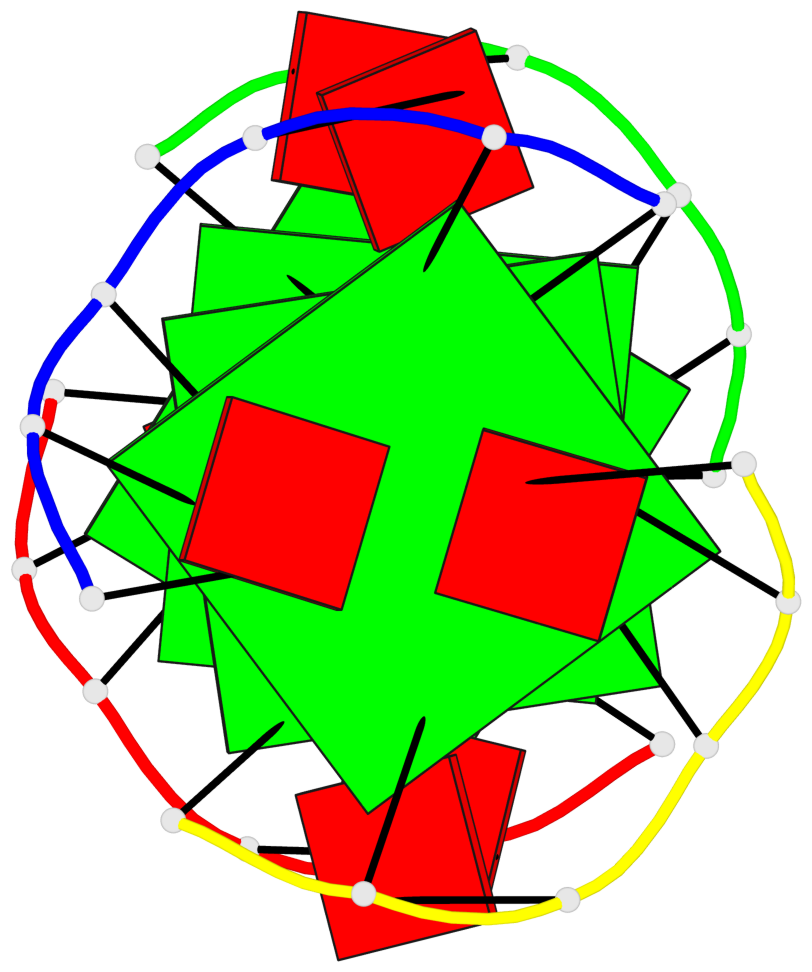
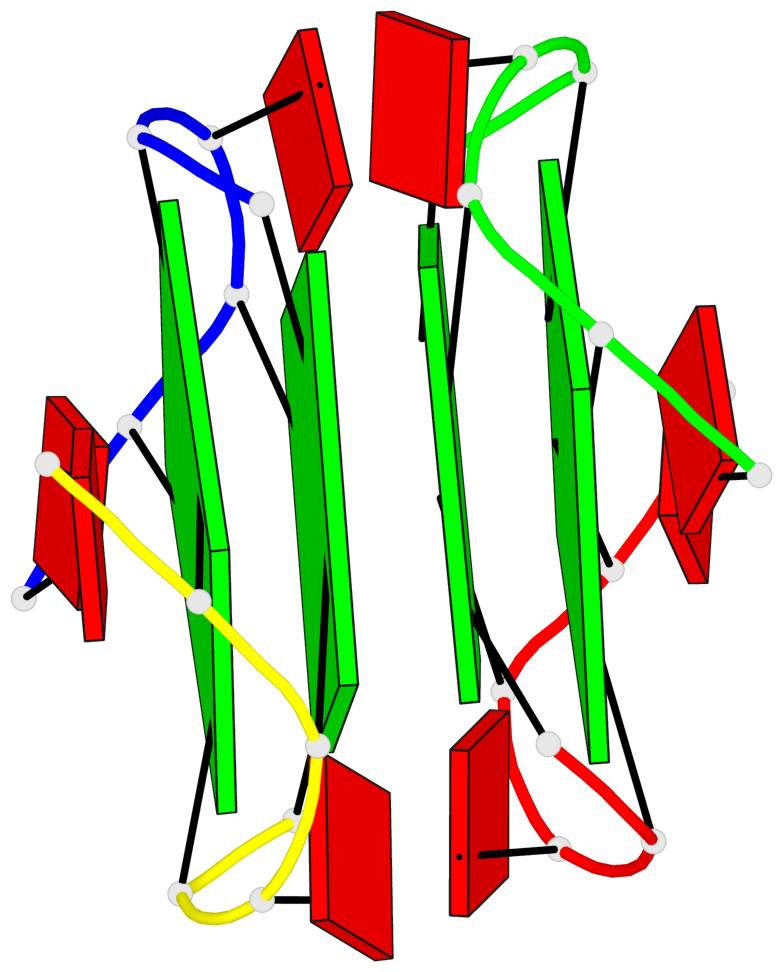
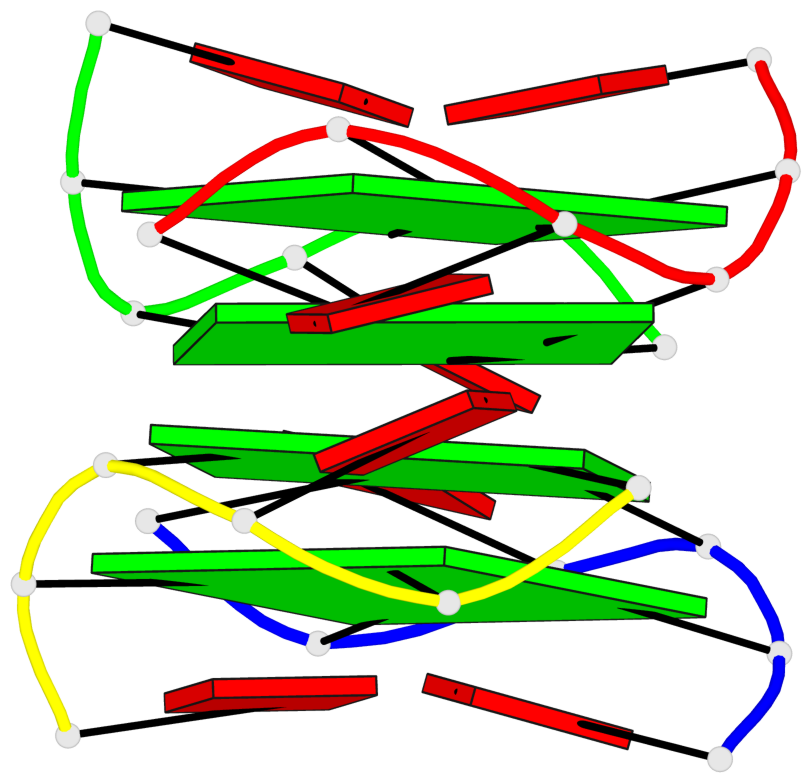
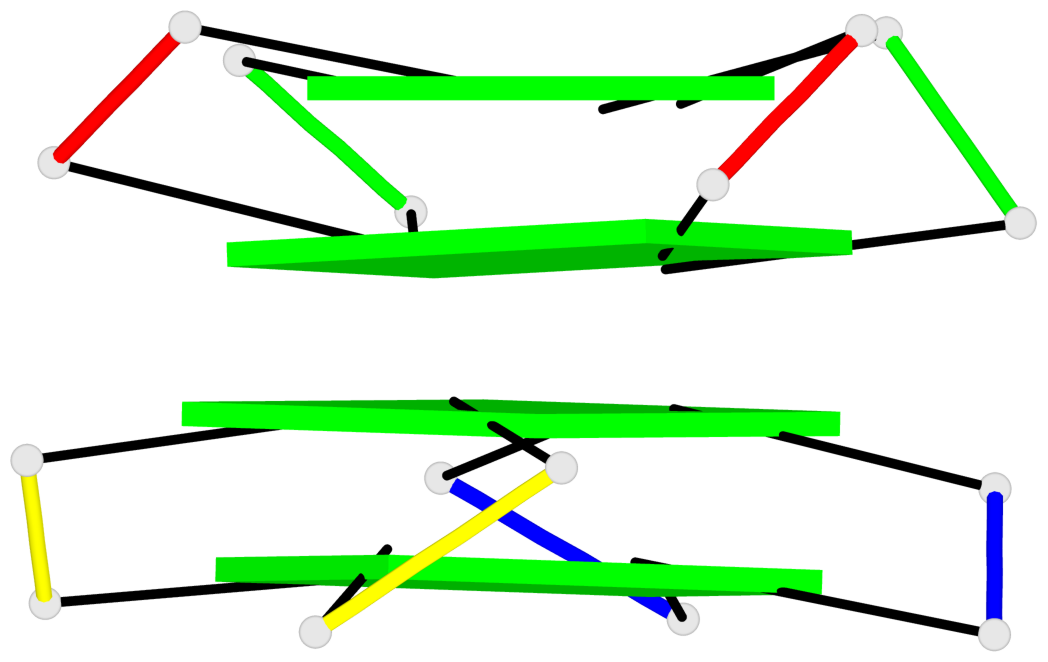
Base-block schematics in six views
List of 4 G-tetrads
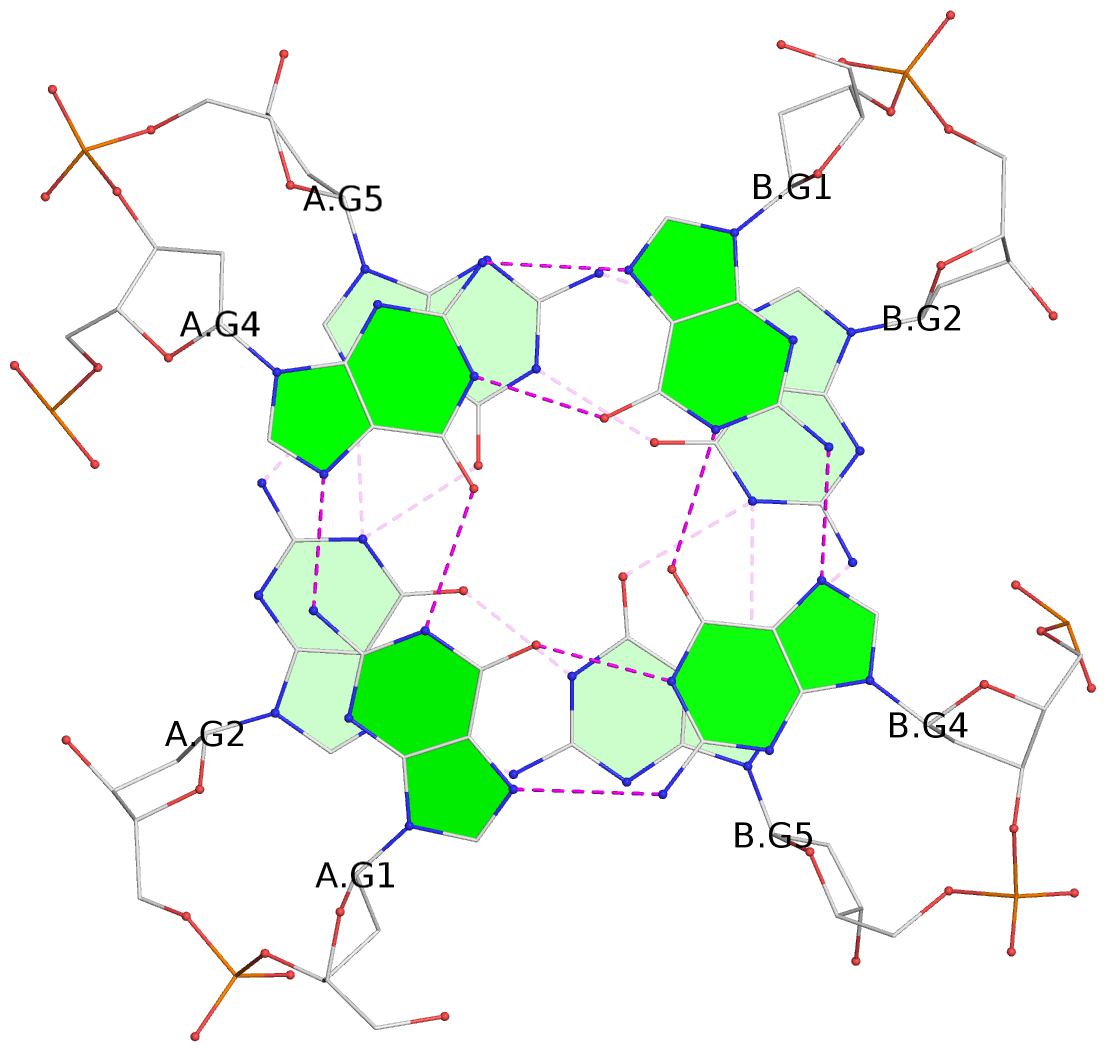
1 glyco-bond=---- sugar=-.-- groove=---- planarity=0.305 type=other nts=4 GGGG A.DG1,A.DG4,B.DG1,B.DG4 2 glyco-bond=---- sugar=-.-. groove=---- planarity=0.211 type=other nts=4 GGGG A.DG2,A.DG5,B.DG2,B.DG5 3 glyco-bond=---- sugar=..-- groove=---- planarity=0.215 type=other nts=4 GGGG C.DG1,C.DG4,D.DG1,D.DG4 4 glyco-bond=---- sugar=---. groove=---- planarity=0.233 type=bowl nts=4 GGGG C.DG2,C.DG5,D.DG2,D.DG5
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 4 G-tetrad layers, inter-molecular, with 2 stems
List of 2 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 2 G-tetrad layers, 2 loops, inter-molecular, UUUU, parallel, parallel(4+0)
Stem#2, 2 G-tetrad layers, 2 loops, inter-molecular, UUUU, parallel, parallel(4+0)
List of 1 G4 coaxial stack
1 G4 helix#1 contains 2 G4 stems: [#1,#2] [5'/5']