Detailed DSSR results for the G-quadruplex: PDB entry 1emq
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 1emq
- Class
- DNA
- Method
- NMR
- Summary
- NMR observation of t-tetrads in a parallel stranded DNA quadruplex formed by saccharomyces cerevisiae telomere repeats
- Reference
- Patel PK, Hosur RV (1999): "NMR observation of T-tetrads in a parallel stranded DNA quadruplex formed by Saccharomyces cerevisiae telomere repeats." Nucleic Acids Res., 27, 2457-2464. doi: 10.1093/nar/27.12.2457.
- Abstract
- We report here the NMR structure of the DNA sequence d-TGGTGGC containing two repeats of Saccharomyces cerevisiae telomere DNA which is unique in that it has a single thymine in the repeat sequence and the number of Gs can vary from one to three. The structure is a novel quadruplex incor-porating T-tetrads formed by symmetrical pairing of four Ts via O4-H3 H-bonds in a plane. This is in contrast to the previous results on other telomeric sequences which contained more than one T in the repeat sequences and they were seen mostly in the flexible regions of the structures. We observed that the T4-tetrad was nicely accommodated in the center of the G-quadruplex, but it caused a small underwinding of the right handed helix. The T tetrad stacked well on the adjacent G3-tetrad, but poorly on the G5 tetrad. Likewise, T1 also formed a stable T-tetrad at the 5' end of the quadruplex. To our knowledge, this is the first report of T-tetrad formation in DNA structures. These observations are of significance from the points of view of both structural diversity and specific recognitions.
- G4 notes
- 4 G-tetrads, 1 G4 helix, 2 G4 stems, 1 G4 coaxial stack, parallel(4+0), UUUU, coaxial interfaces: 3'/5'
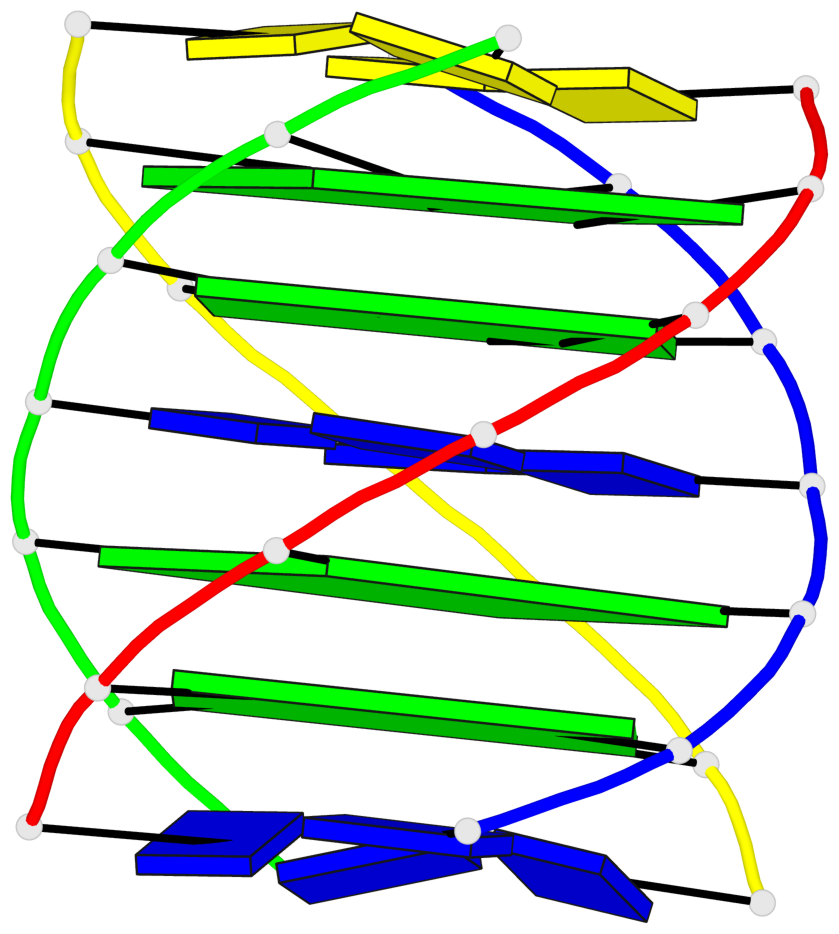
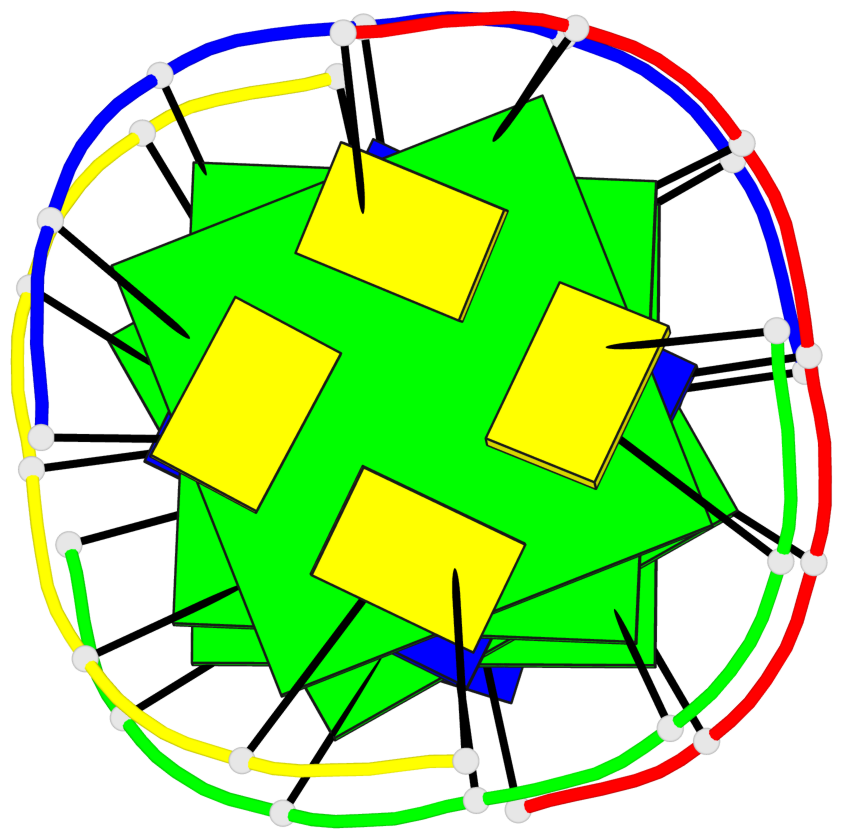
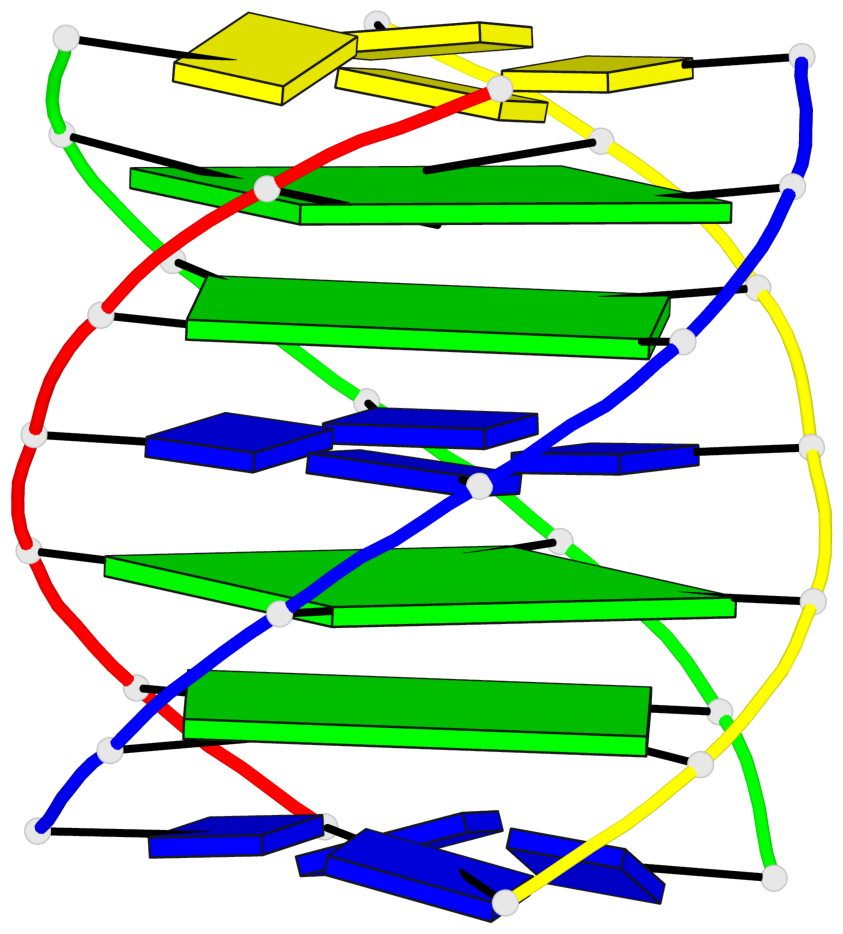
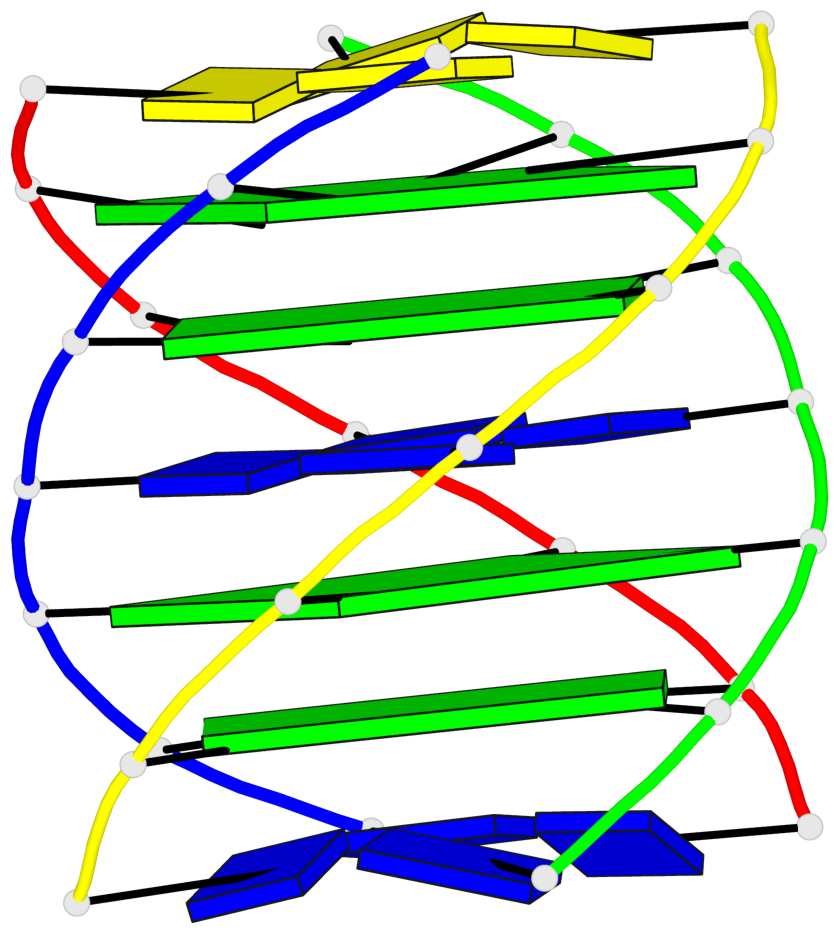
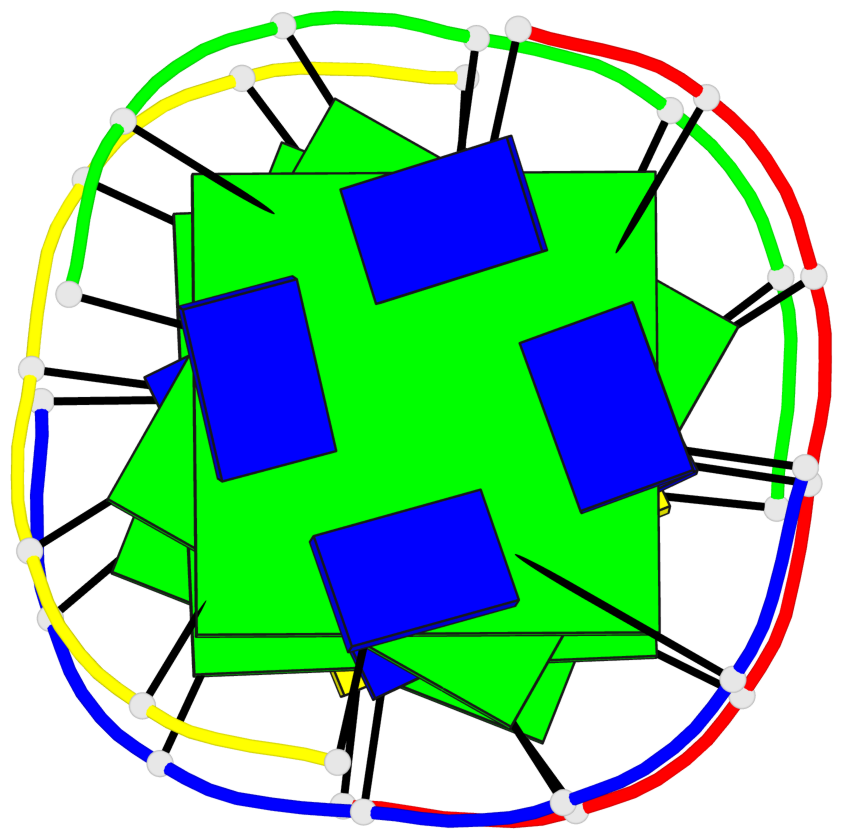
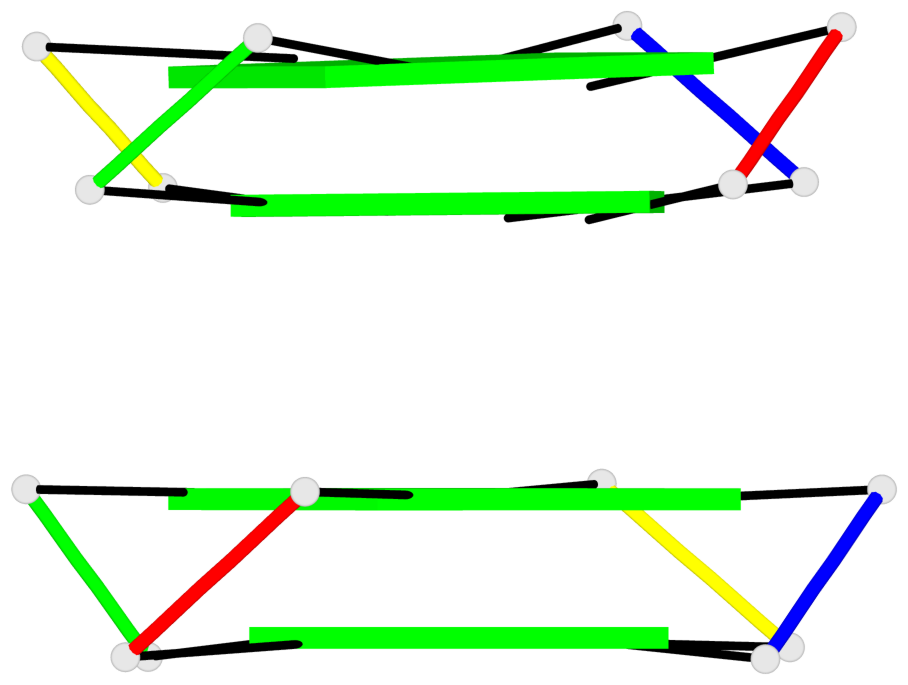
Base-block schematics in six views
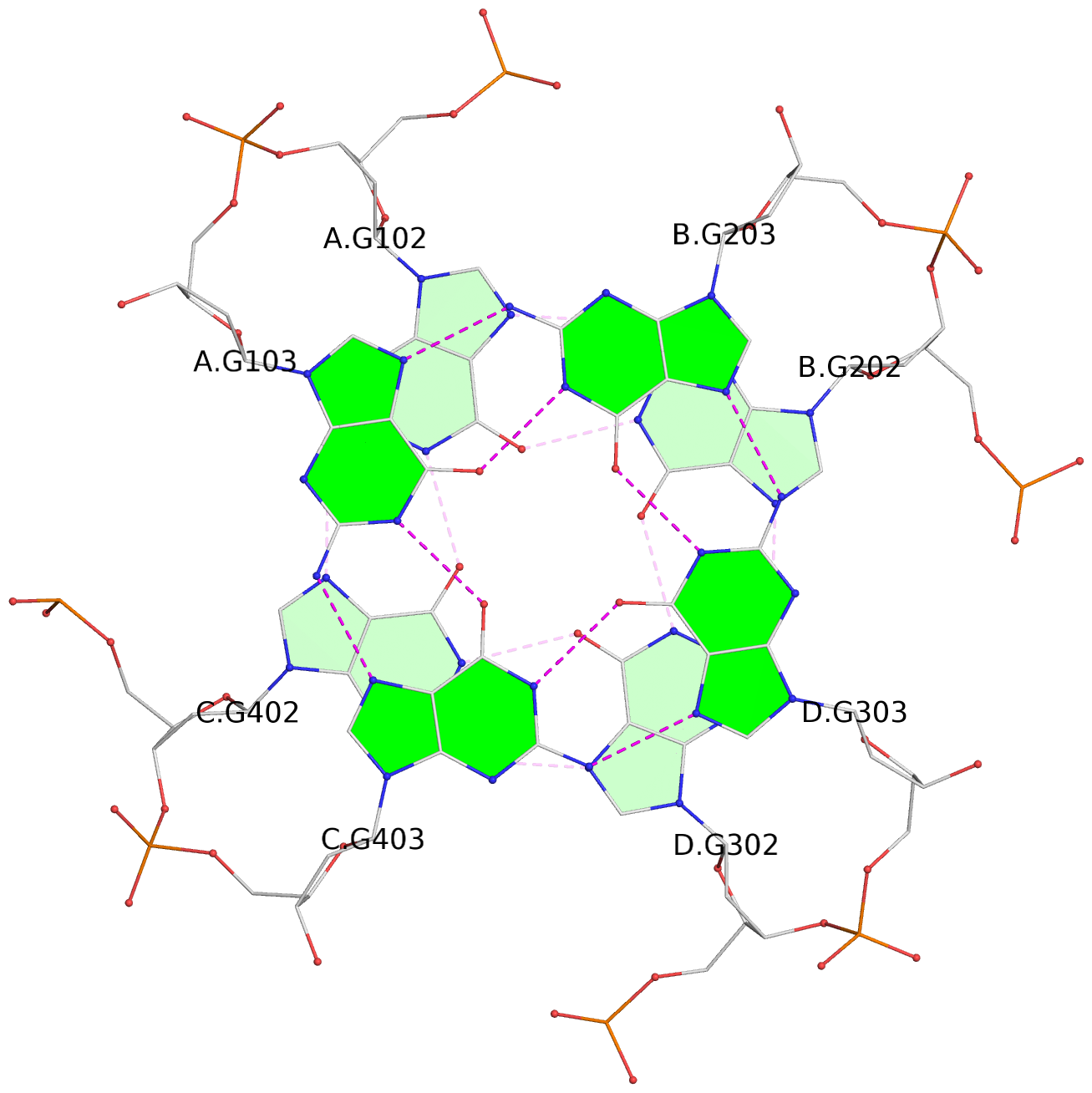
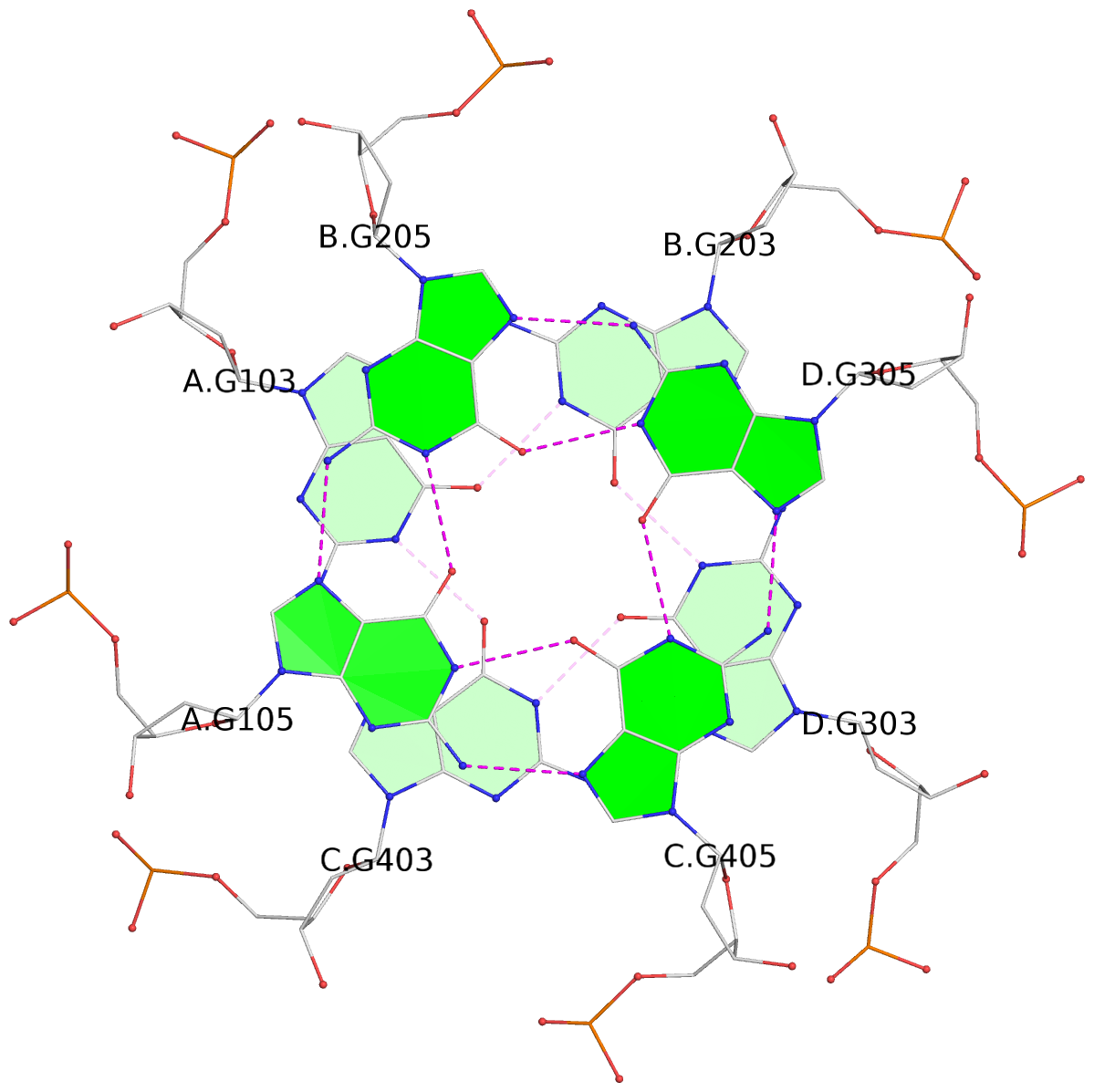
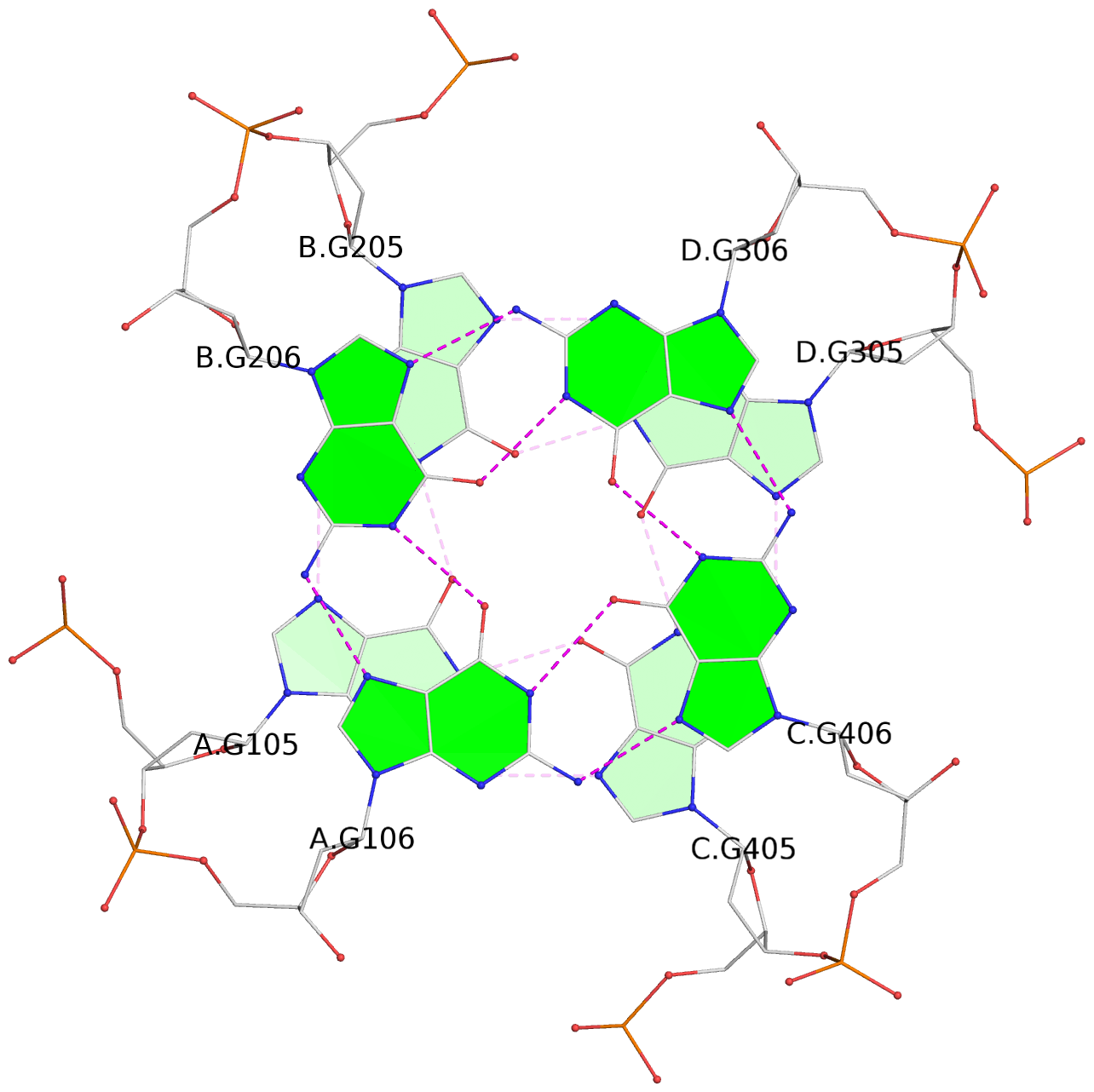
List of 4 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.147 type=planar nts=4 GGGG A.DG102,C.DG402,D.DG302,B.DG202 2 glyco-bond=---- sugar=---- groove=---- planarity=0.070 type=planar nts=4 GGGG A.DG103,C.DG403,D.DG303,B.DG203 3 glyco-bond=---- sugar=---- groove=---- planarity=0.192 type=bowl nts=4 GGGG A.DG105,C.DG405,D.DG305,B.DG205 4 glyco-bond=---- sugar=---- groove=---- planarity=0.194 type=other nts=4 GGGG A.DG106,C.DG406,D.DG306,B.DG206
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 4 G-tetrad layers, inter-molecular, with 2 stems
List of 2 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 2 G-tetrad layers, 0 loops, inter-molecular, UUUU, parallel, parallel(4+0)
Stem#2, 2 G-tetrad layers, 0 loops, inter-molecular, UUUU, parallel, parallel(4+0)
List of 1 G4 coaxial stack
1 G4 helix#1 contains 2 G4 stems: [#1,#2] [3'/5']