Detailed DSSR results for the G-quadruplex: PDB entry 1evo
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 1evo
- Class
- DNA
- Method
- NMR
- Summary
- NMR observation of a novel c-tetrad
- Reference
- Patel PK, Bhavesh NS, Hosur RV (2000): "NMR observation of a novel C-tetrad in the structure of the SV40 repeat sequence GGGCGG." Biochem.Biophys.Res.Commun., 270, 967-971. doi: 10.1006/bbrc.2000.2479.
- Abstract
- We report the NMR structure of the DNA sequence d-TGGGCGGT in Na(+) solutions at neutral pH, containing a repeat sequence from SV40 viral genome. The structure is a novel quadruplex incorporating the C-tetrad formed by symmetrical pairing of four Cs via NH(2)&bond;O(2) H-bonds in a plane. The C-tetrad has a wider cavity compared to G-tetrads and stacks well over the adjacent G4-tetrad, but poorly on the G6 tetrad. The quadruplex helix is largely underwound by 8-10 degrees compared to B-DNA except at the C5-G6 step. To our knowledge this is the first report of C-tetrad formation in DNA structures, and would be of significance from the point of view of both structural diversity and specific recognition.
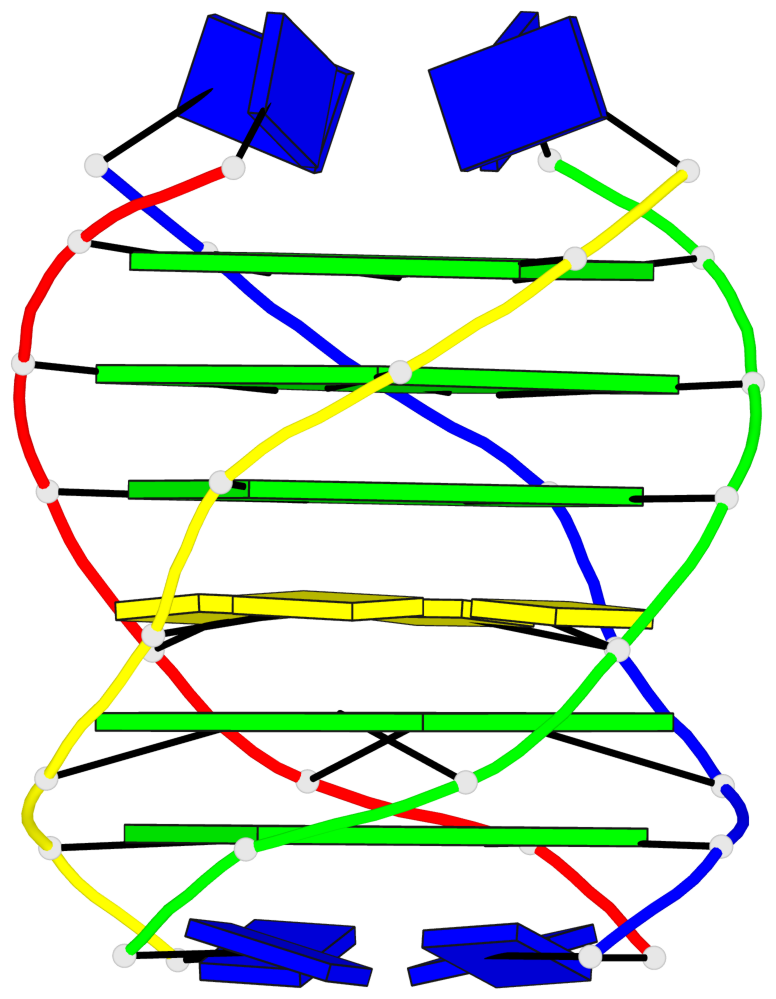
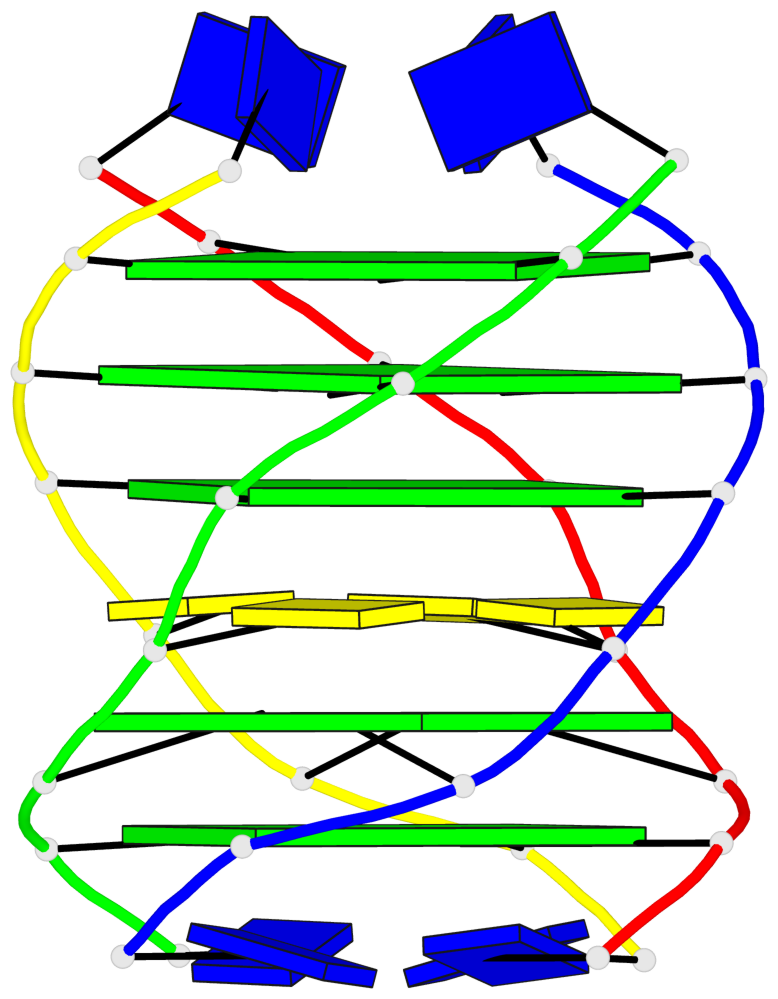
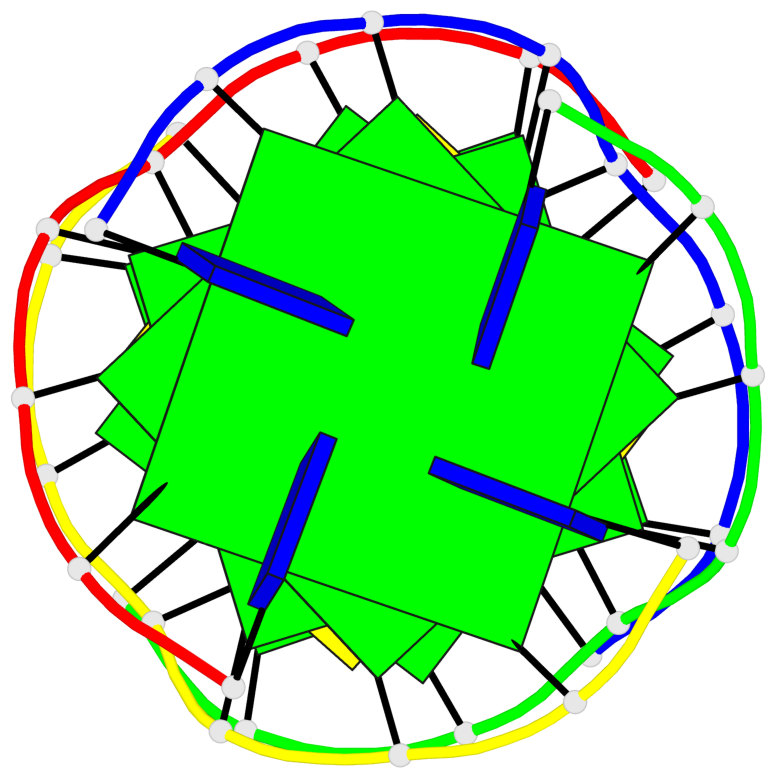
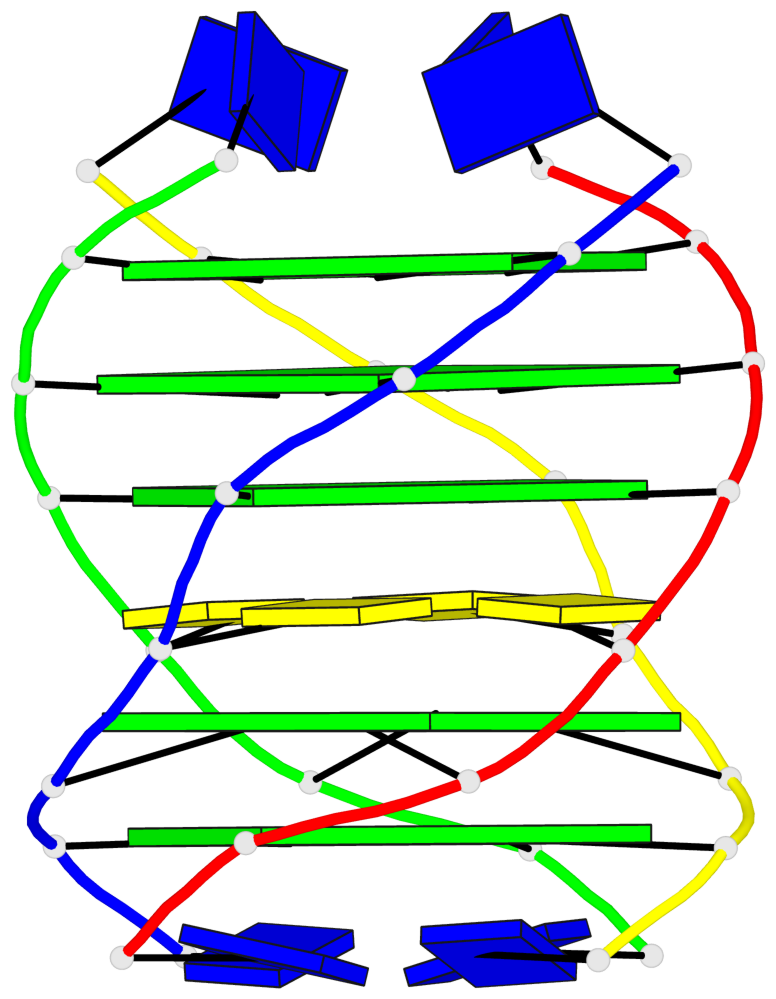
- G4 notes
- 5 G-tetrads, 1 G4 helix, 2 G4 stems, 1 G4 coaxial stack, parallel(4+0), UUUU, coaxial interfaces: 3'/5'
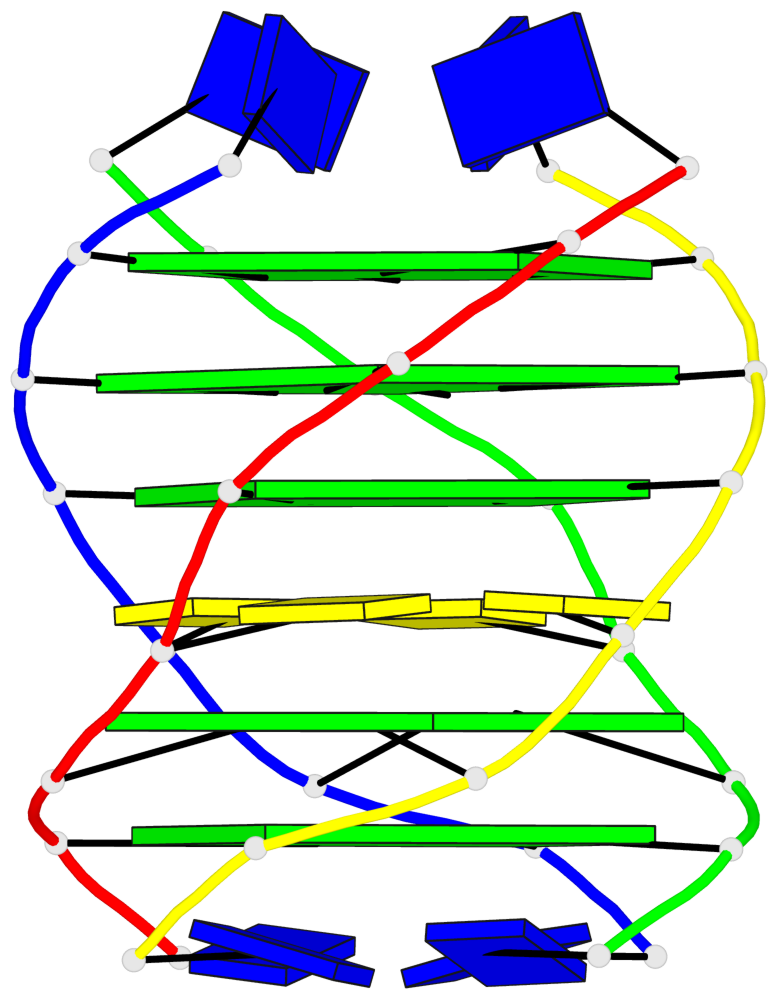
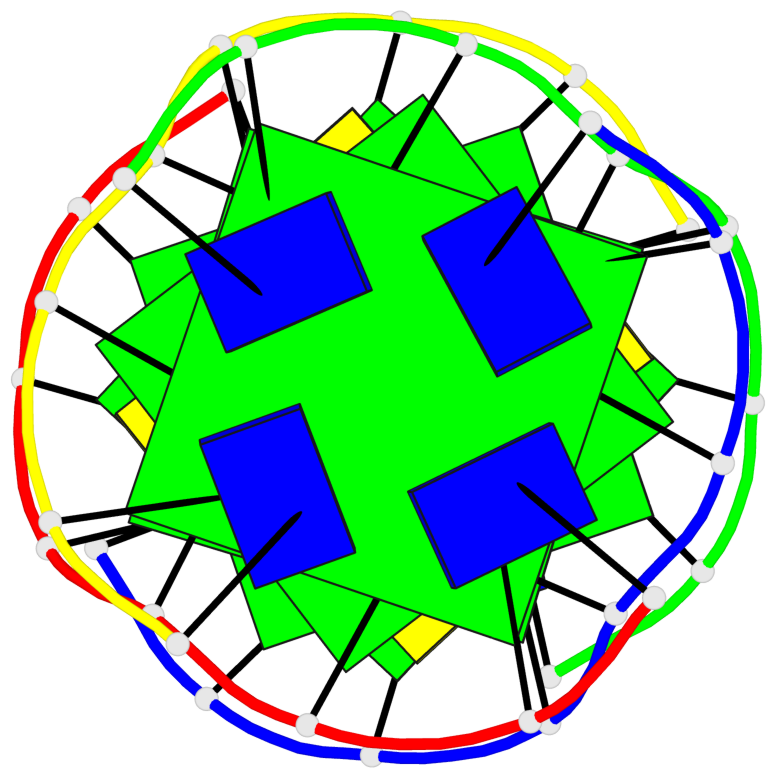
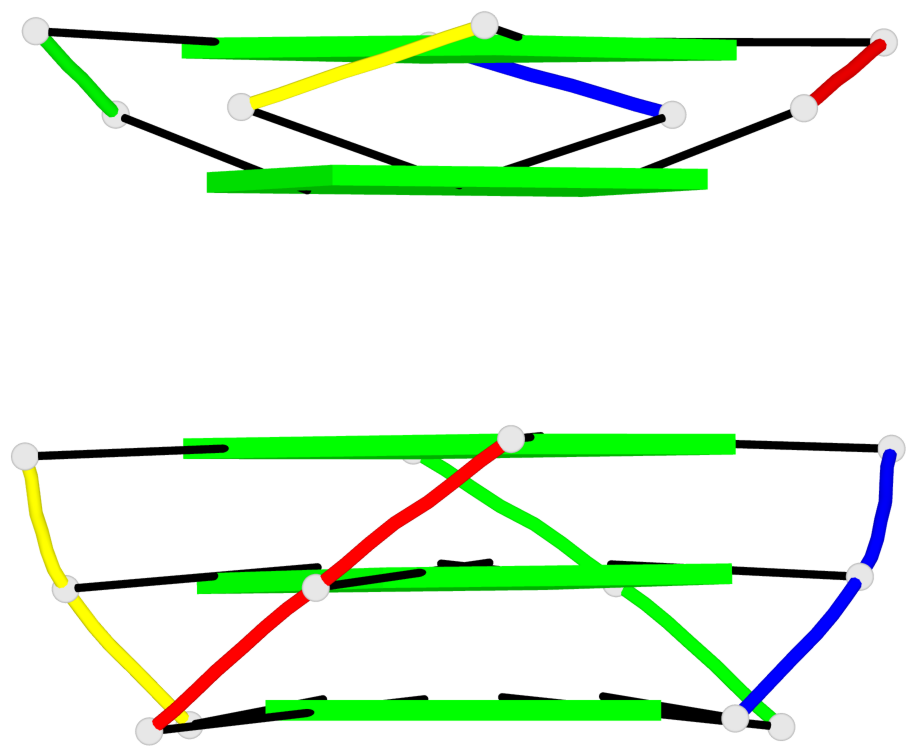
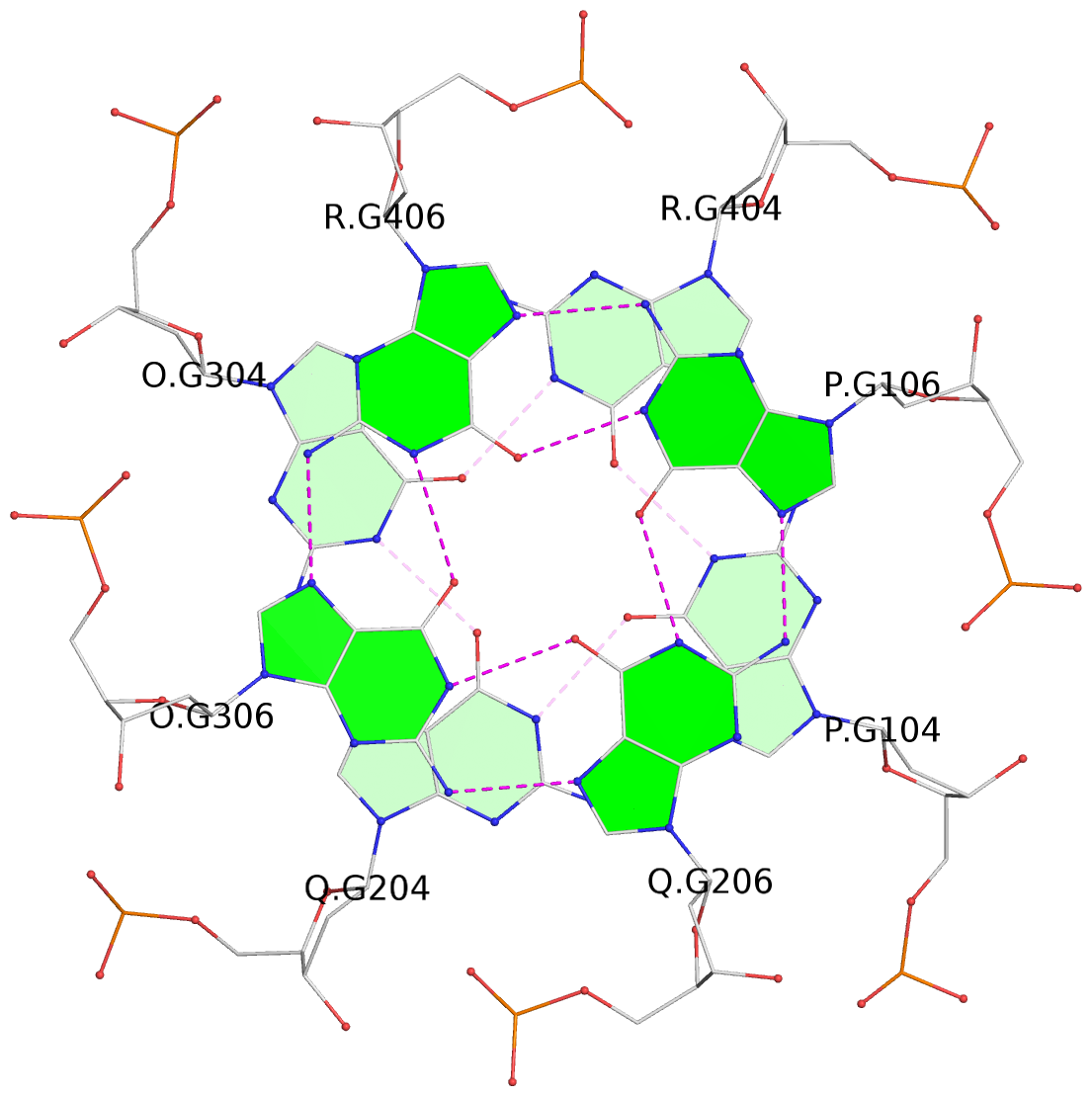
Base-block schematics in six views
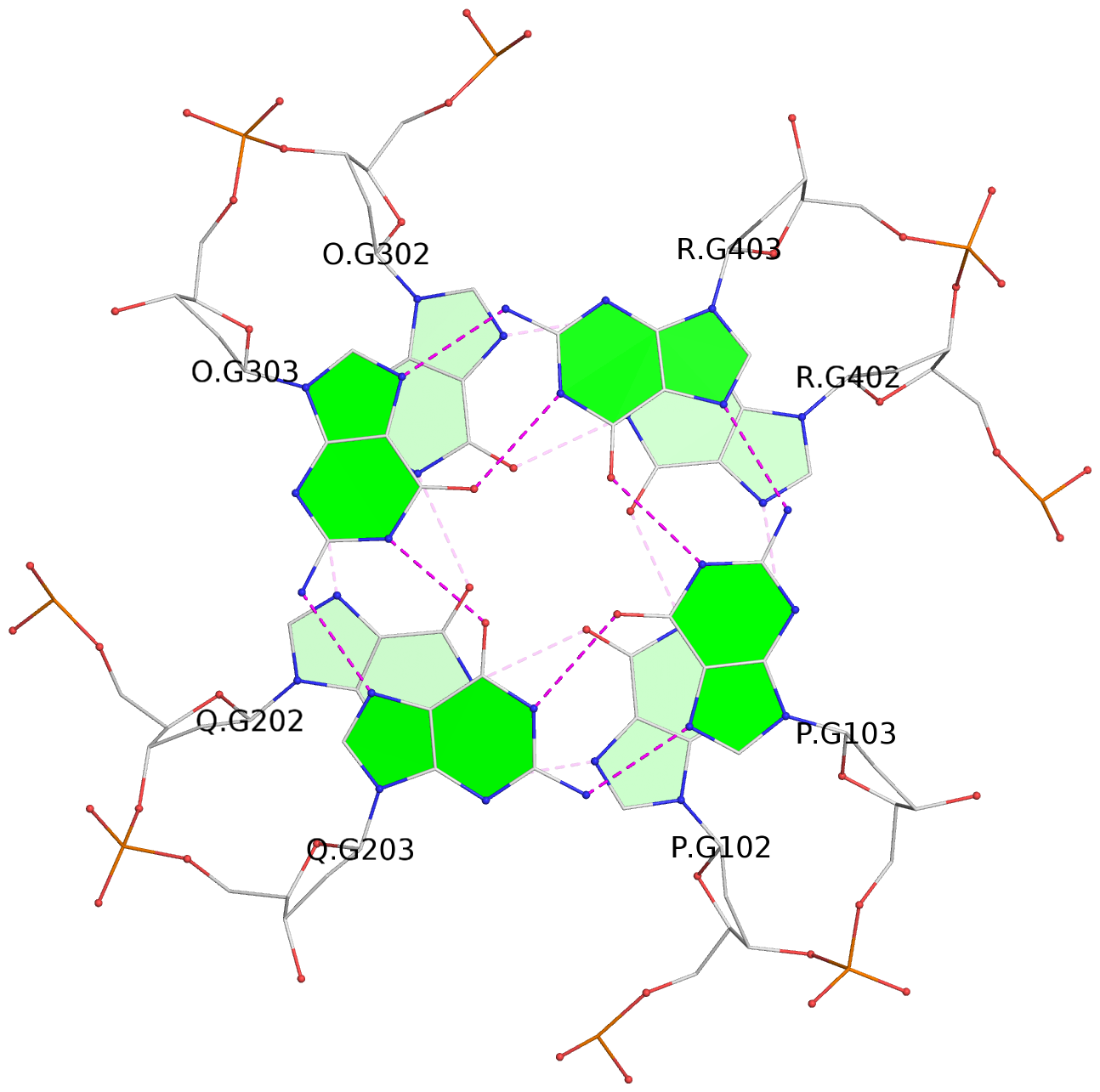
List of 5 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.409 type=other nts=4 GGGG O.DG302,Q.DG202,P.DG102,R.DG402 2 glyco-bond=---- sugar=.-.. groove=---- planarity=0.316 type=other nts=4 GGGG O.DG303,Q.DG203,P.DG103,R.DG403 3 glyco-bond=---- sugar=---- groove=---- planarity=0.190 type=other nts=4 GGGG O.DG304,Q.DG204,P.DG104,R.DG404 4 glyco-bond=---- sugar=---- groove=---- planarity=0.235 type=other nts=4 GGGG O.DG306,Q.DG206,P.DG106,R.DG406 5 glyco-bond=---- sugar=---- groove=---- planarity=0.210 type=other nts=4 GGGG O.DG307,Q.DG207,P.DG107,R.DG407
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 5 G-tetrad layers, inter-molecular, with 2 stems
List of 2 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 3 G-tetrad layers, 0 loops, inter-molecular, UUUU, parallel, parallel(4+0)
Stem#2, 2 G-tetrad layers, 0 loops, inter-molecular, UUUU, parallel, parallel(4+0)
List of 1 G4 coaxial stack
1 G4 helix#1 contains 2 G4 stems: [#1,#2] [3'/5']