Detailed DSSR results for the G-quadruplex: PDB entry 1f3s
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 1f3s
- Class
- DNA
- Method
- NMR
- Summary
- Solution structure of DNA sequence gggttcagg forms gggg tetrade and g(c-a) triad.
- Reference
- Kettani A, Basu G, Gorin A, Majumdar A, Skripkin E, Patel DJ (2000): "A two-stranded template-based approach to G.(C-A) triad formation: designing novel structural elements into an existing DNA framework." J.Mol.Biol., 301, 129-146. doi: 10.1006/jmbi.2000.3932.
- Abstract
- We have designed a DNA sequence, d(G-G-G-T-T-C-A-G-G), which dimerizes to form a 2-fold symmetric G-quadruplex in which G(syn). G(anti).G(syn).G(anti) tetrads are sandwiched between all trans G. (C-A) triads. The NMR-based solution structural analysis was greatly aided by monitoring hydrogen bond alignments across N-H...N and N-H...O==C hydrogen bonds within the triad and tetrad, in a uniformly ((13)C,(15)N)-labeled sample of the d(G-G-G-T-T-C-A-G-G) sequence. The solution structure establishes that the guanine base-pairs with the cytosine through Watson-Crick G.C pair formation and with adenine through sheared G.A mismatch formation within the G.(C-A) triad. A model of triad DNA was constructed that contains the experimentally determined G.(C-A) triad alignment as the repeating stacked unit.
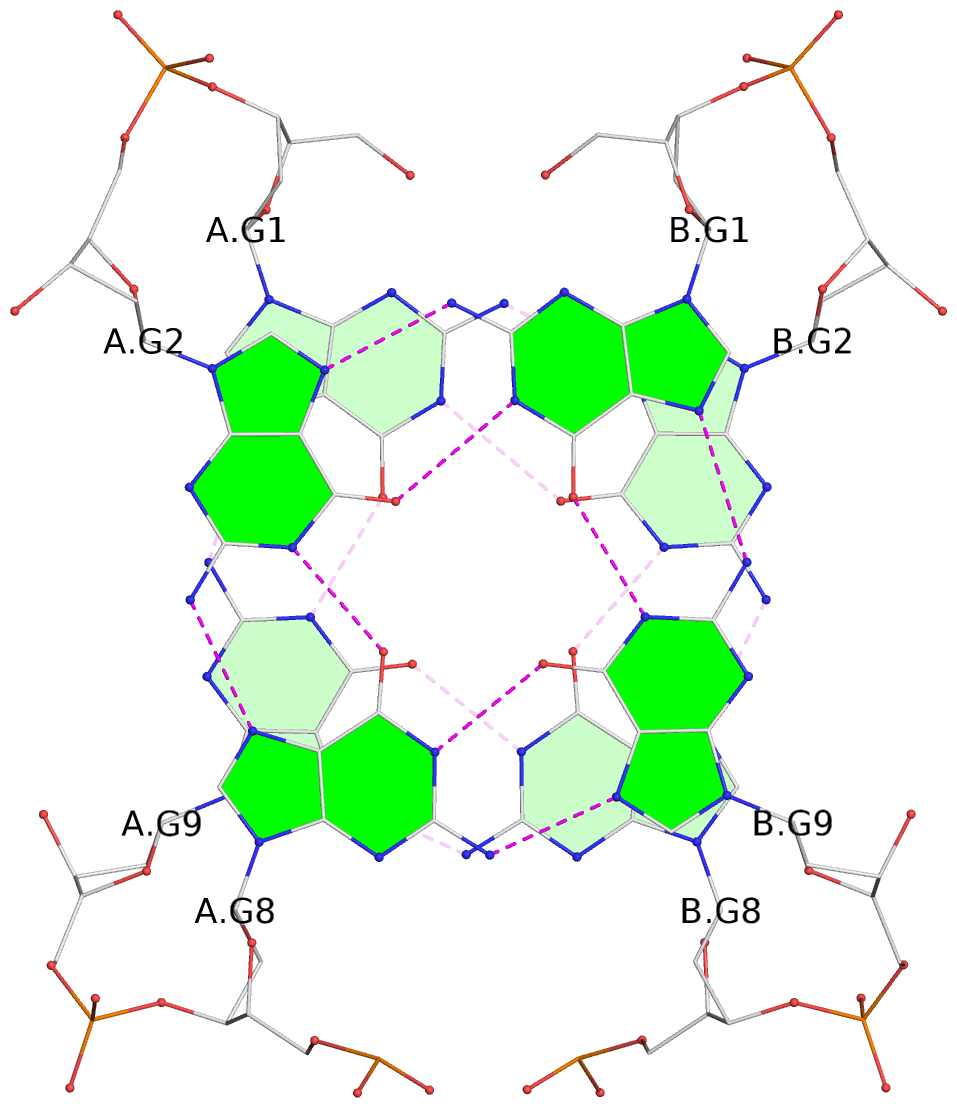
- G4 notes
- 2 G-tetrads, 1 G4 helix, 1 G4 stem, (2+2), UDUD
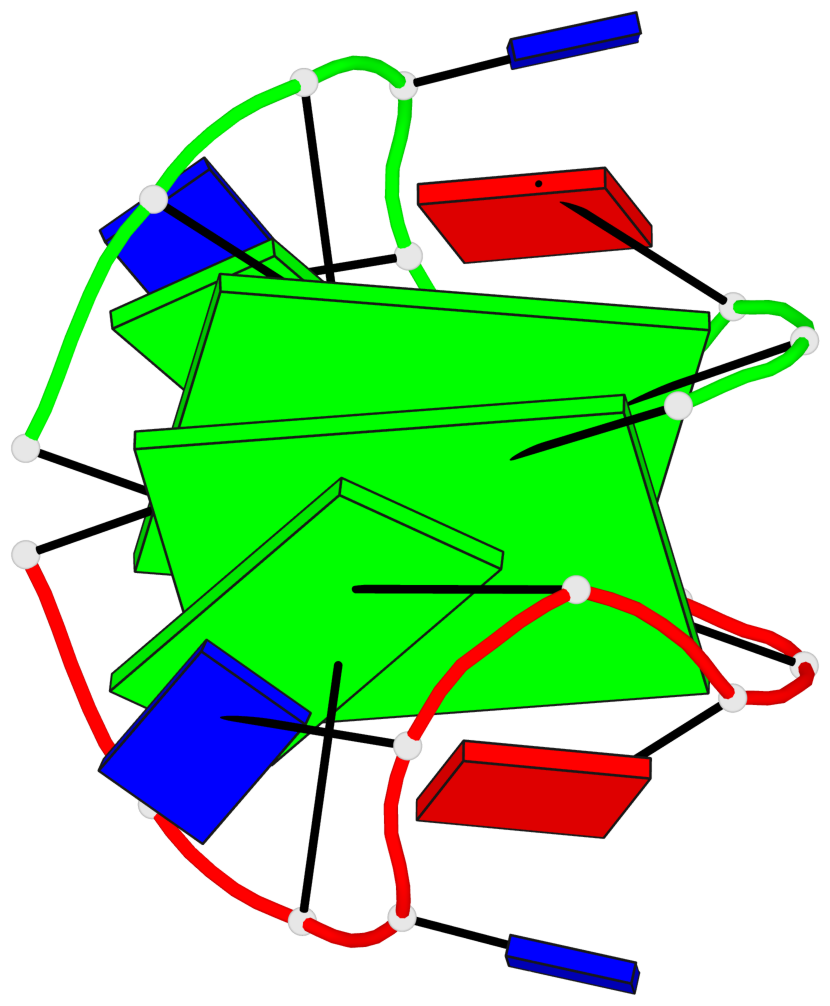
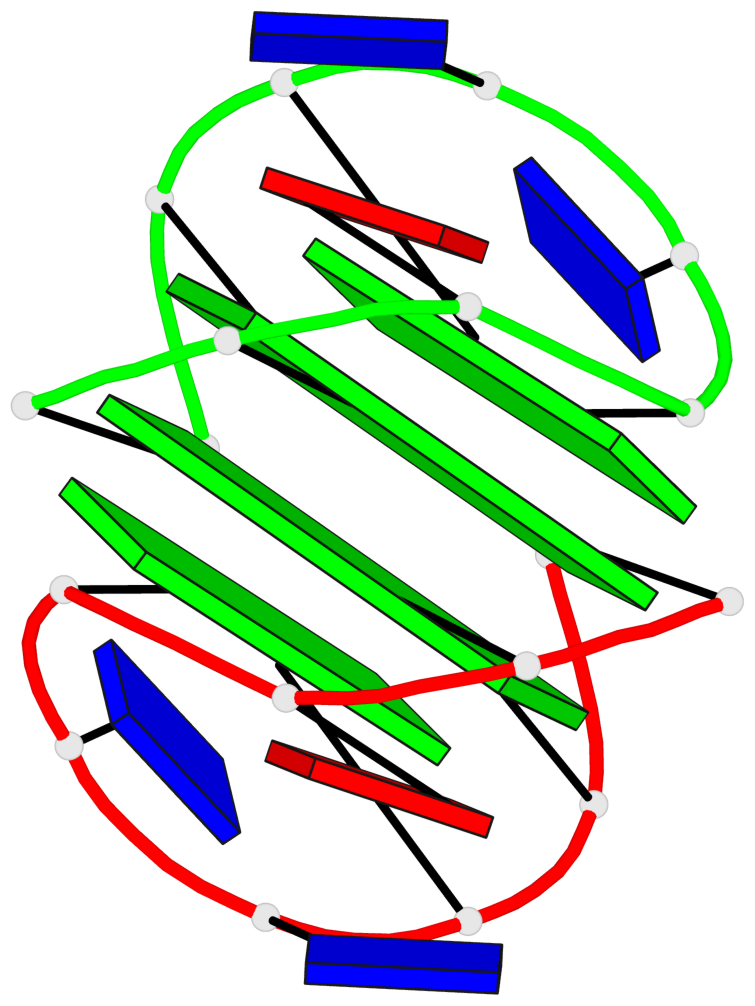
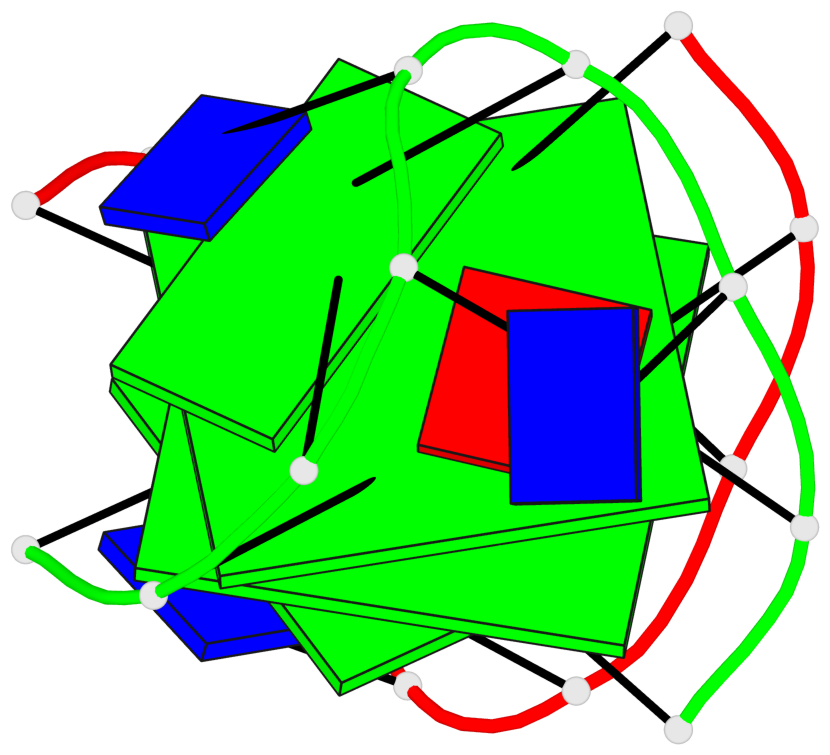
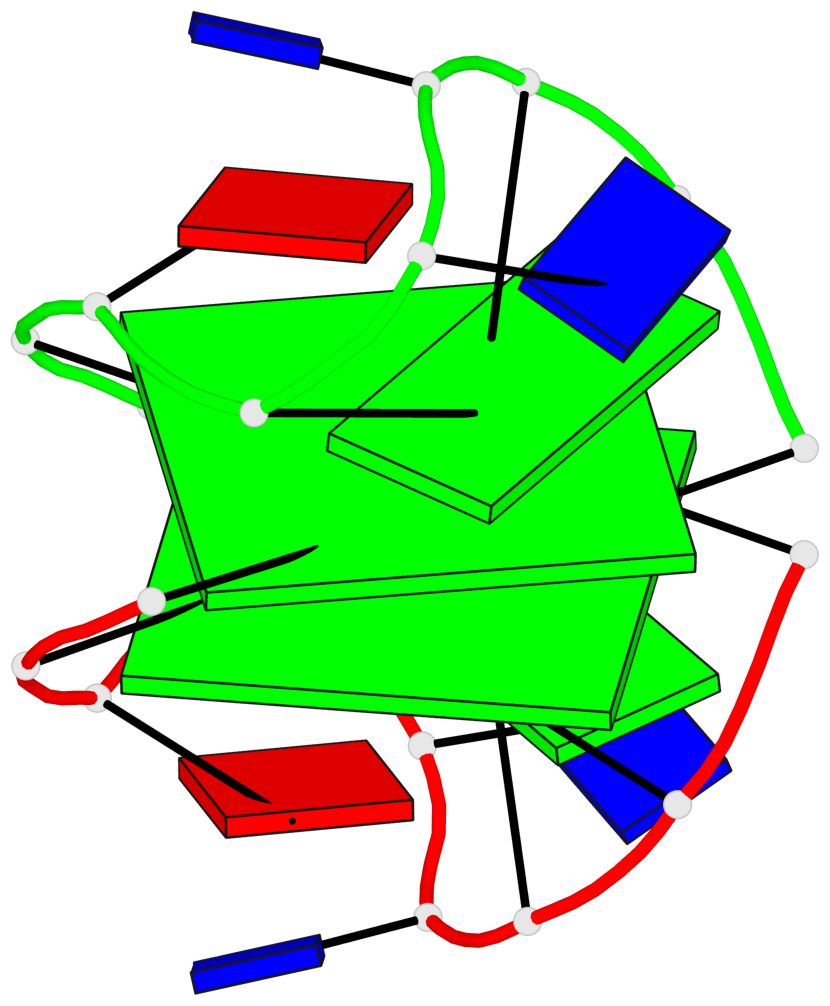
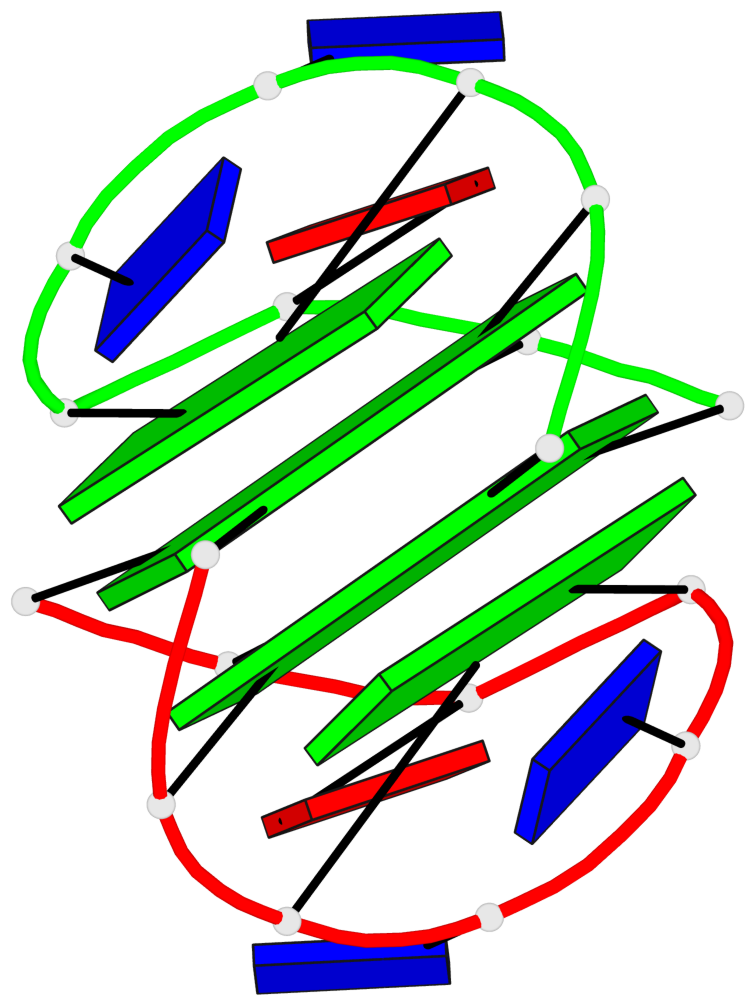
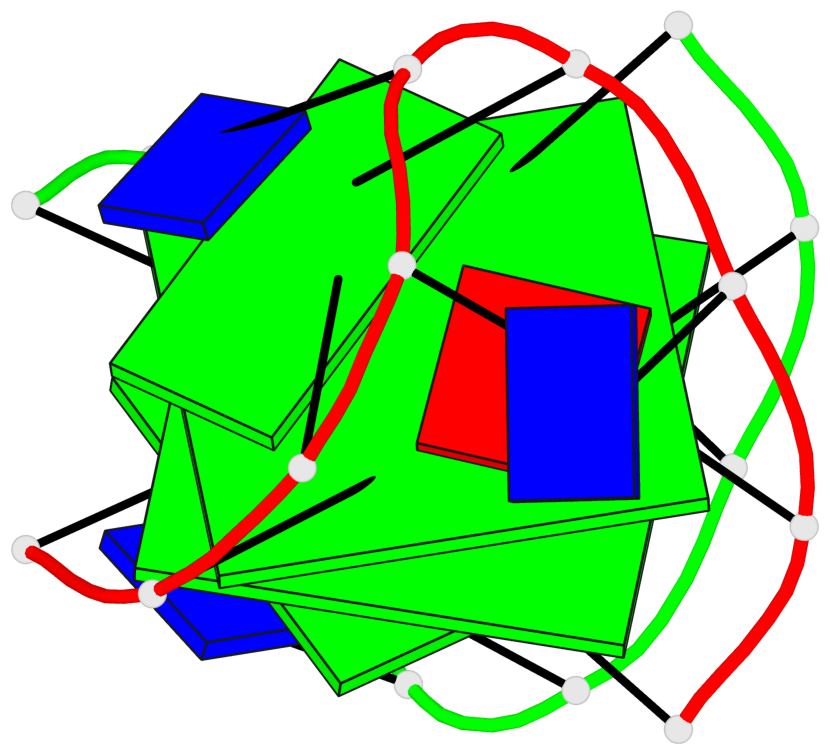
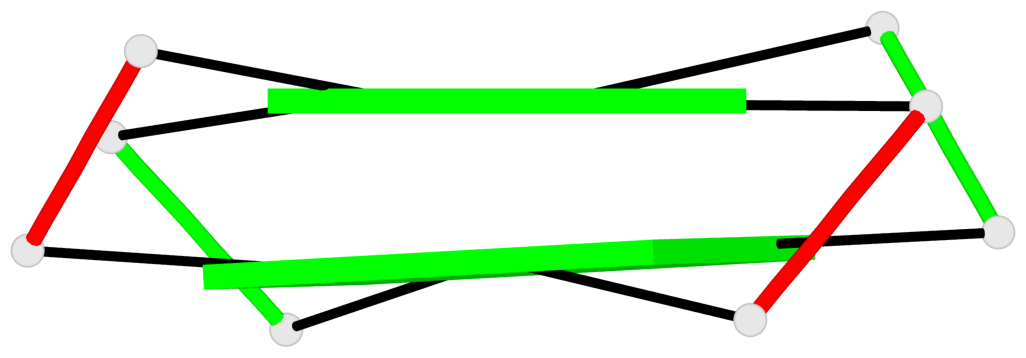
Base-block schematics in six views
List of 2 G-tetrads
1 glyco-bond=s-s- sugar=---- groove=wnwn planarity=0.138 type=planar nts=4 GGGG A.DG1,A.DG9,B.DG8,B.DG2 2 glyco-bond=-s-s sugar=---- groove=wnwn planarity=0.141 type=planar nts=4 GGGG A.DG2,A.DG8,B.DG9,B.DG1
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 2 G-tetrad layers, inter-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.