Detailed DSSR results for the G-quadruplex: PDB entry 1fqp
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 1fqp
- Class
- DNA
- Method
- NMR
- Summary
- Intramolecular quadruplex DNA with three gggg repeats, NMR, ph 6.7, 0.1 m na+ and 4 mm (strand concentration), 5 structures
- Reference
- Keniry MA, Strahan GD, Owen EA, Shafer RH (1995): "Solution structure of the Na+ form of the dimeric guanine quadruplex [d(G3T4G3)]2." Eur.J.Biochem., 233, 631-643. doi: 10.1111/j.1432-1033.1995.631_2.x.
- Abstract
- The solution structure of the DNA quadruplex formed by the association of two strands of the DNA oligonucleotide, d(G3T4G3), in NaCl solution has been determined by 1H two-dimensional NMR techniques, full relaxation matrix calculations and restrained molecular dynamics. The refined structure incorporates the sequences 5'-G1sG2AG3AT4AT5AT6AT7AG8sG9AG10A-3' and 5'-G11sG12AG13AT14AT15AT16AT17AG18sG19sG20A-3' (where S and A denote syn and anti, respectively) in a three-quartet, diagonal-looped structure that we [Strahan, G. D., Shafer, R. H. & Keniry, M. A. (1994) Nucleic Acids Res. 22, 5447-5455] and others [Smith, F. W., Lau, F. W. & Feigon, J. (1994) Proc. Natl. Acad. Sci. USA 91, 10546-10550] have described. The loop structure is compact and incorporates many of the features found in duplex hairpin loops including base stacking, intraloop hydrogen bonding and extensive van der Waals' interactions. The first and third loop thymines stack over the outermost G-quartet and are also associated by hydrogen bonding. The second and the fourth loop thymines fold inwards in order to enhance van der Waals' interactions. The unexpected sequential syn-syn deoxyguanosines in the quadruplex stem appear to be a direct consequence of the way DNA oligonucleotides fold and the subsequent search for the most stable loop structure. The implications of loop sequence and length on the structure of quadruplexes are discussed.
- G4 notes
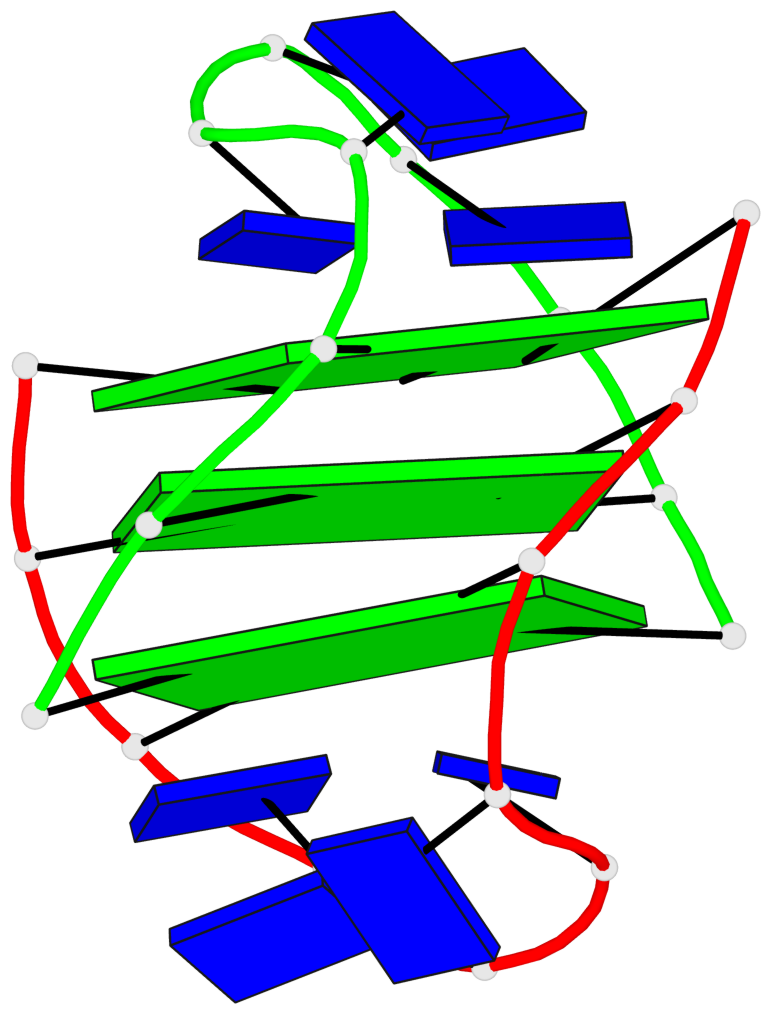
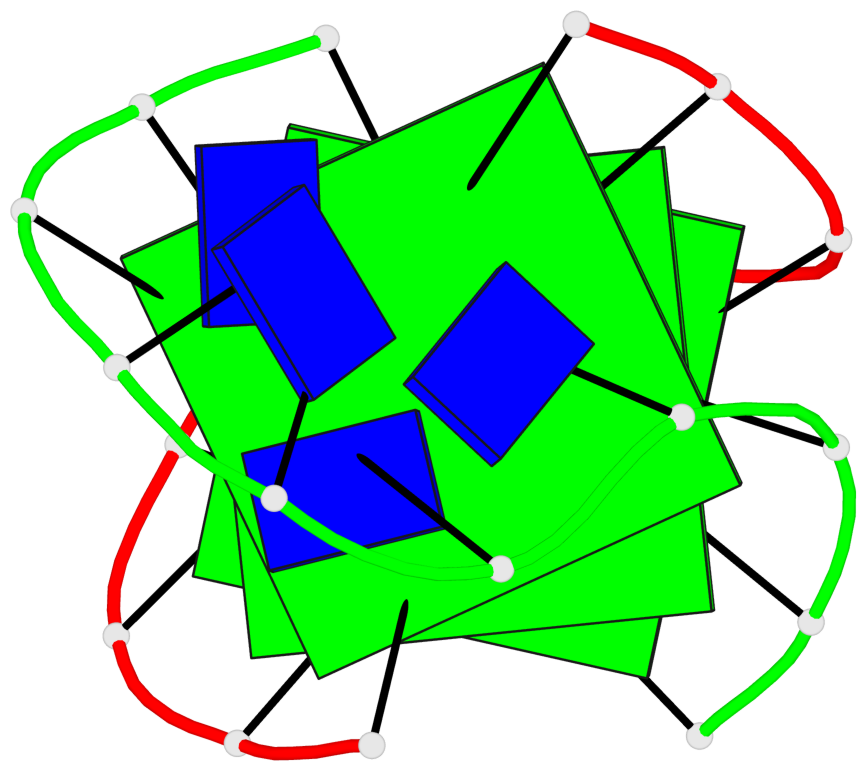
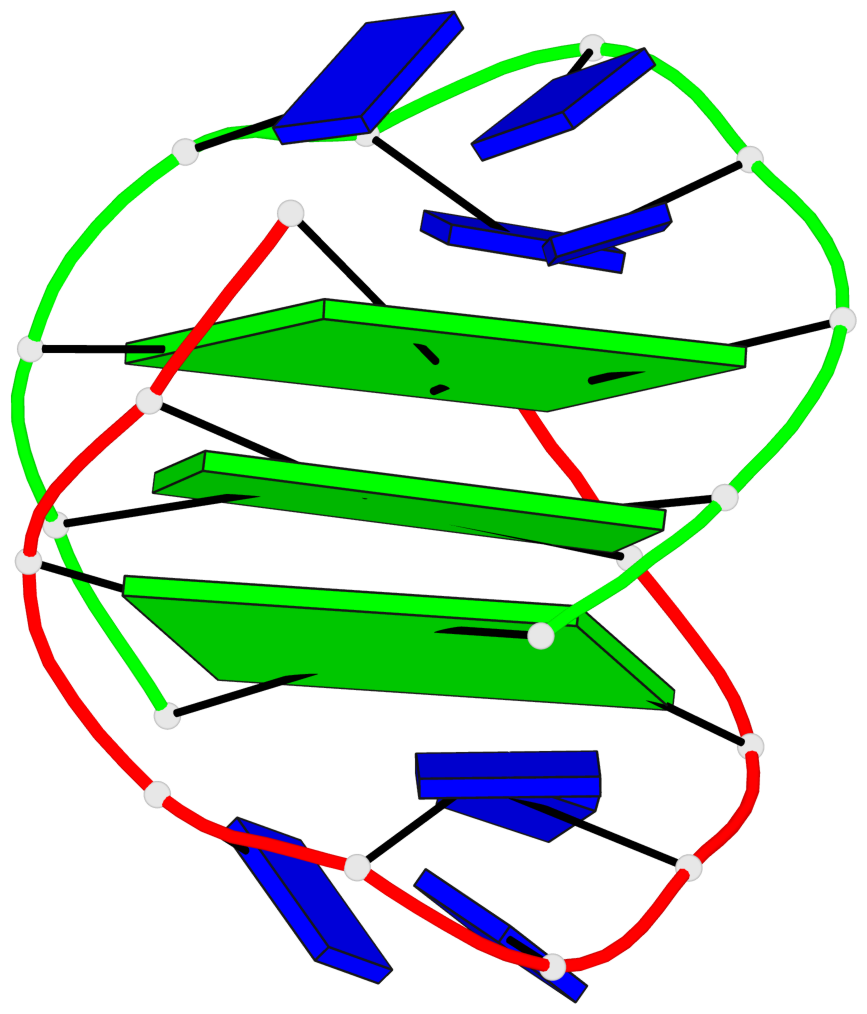
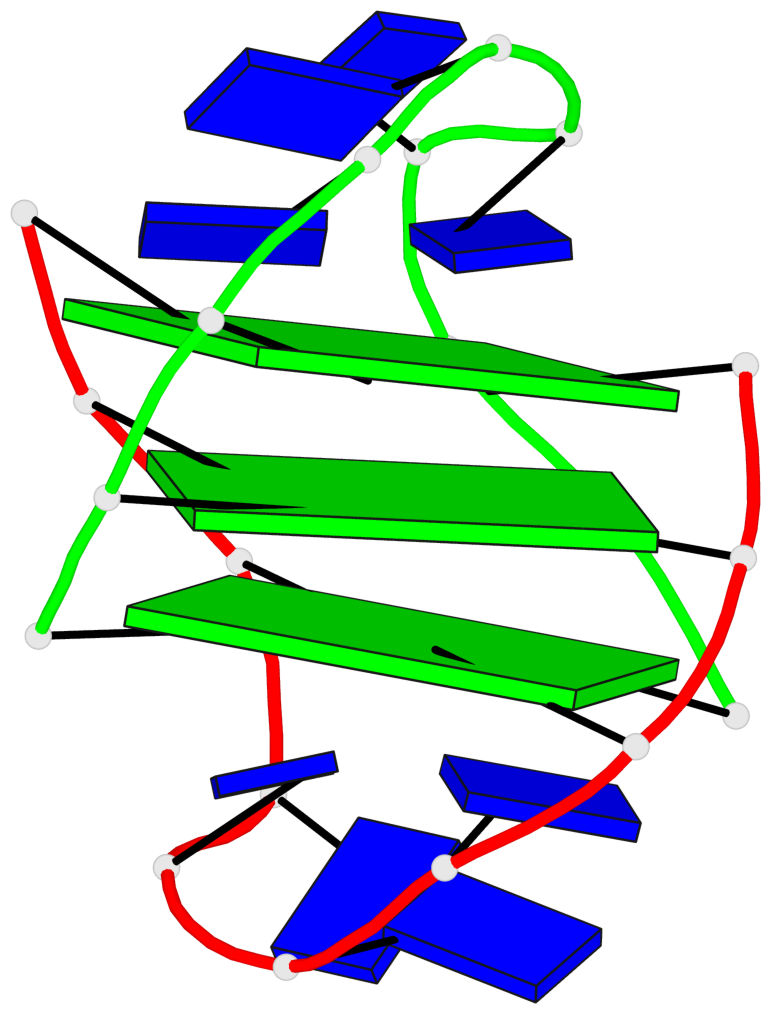
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, (2+2), UDDU
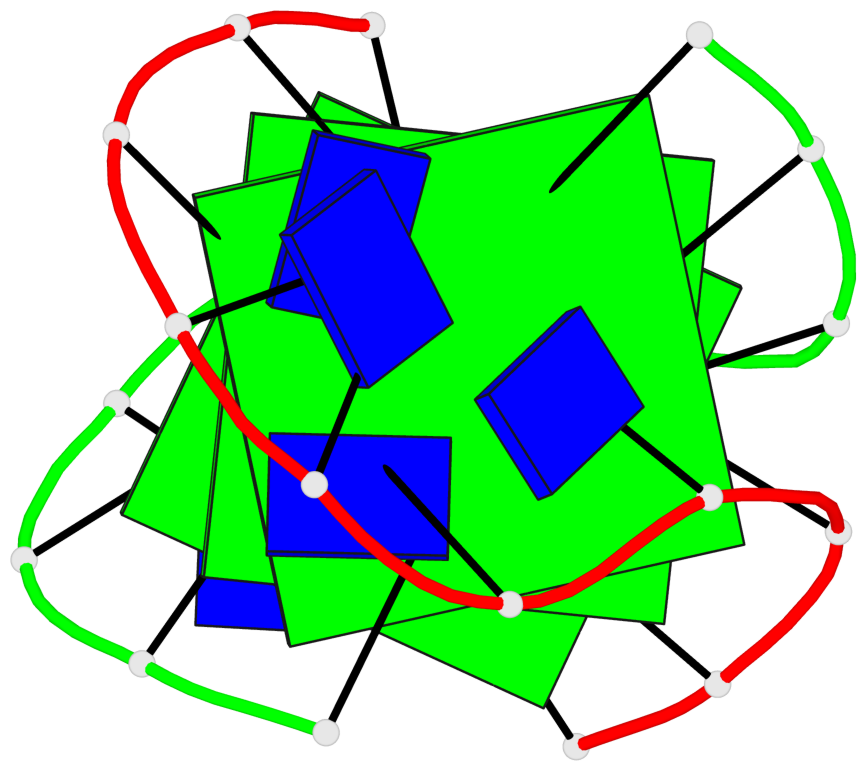
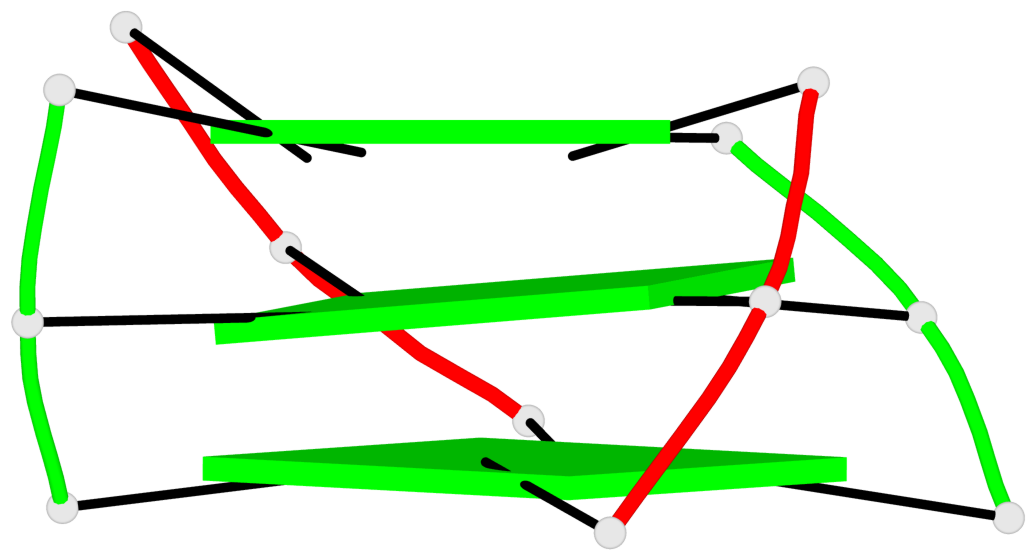
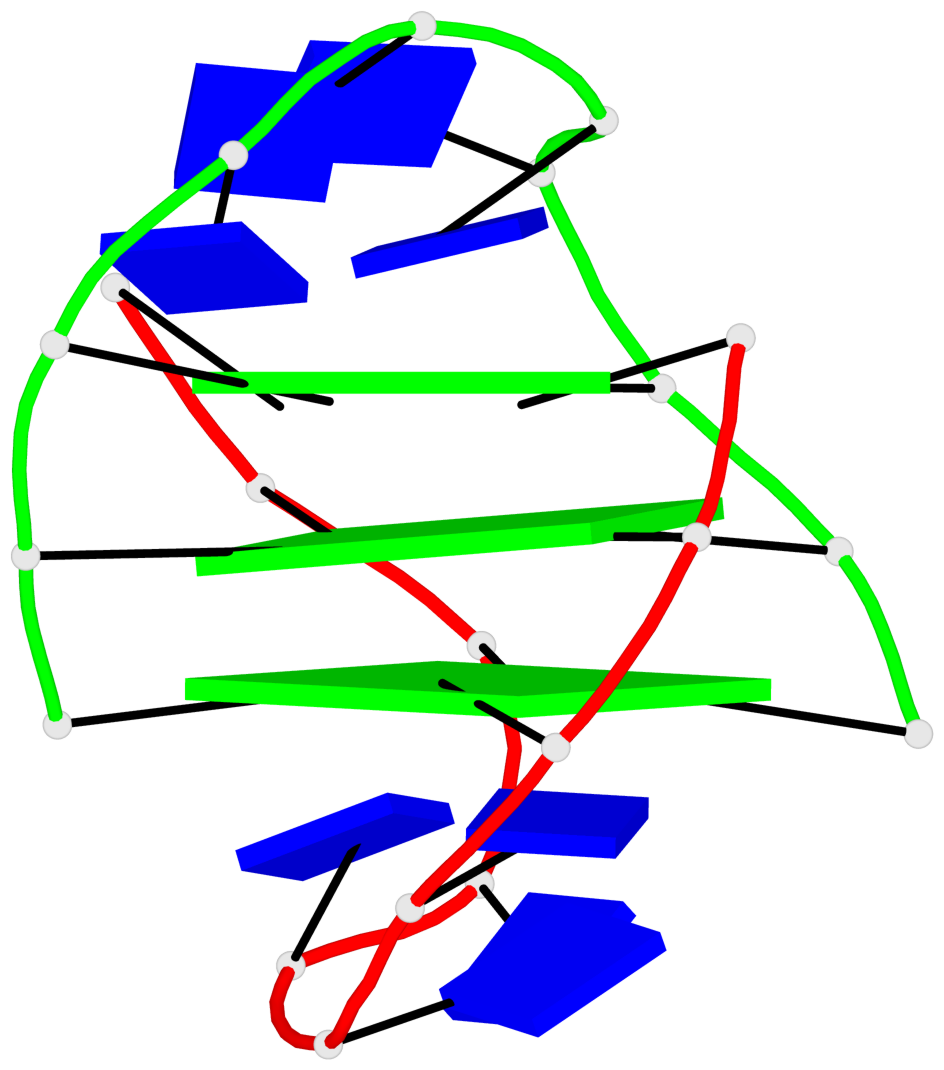
Base-block schematics in six views
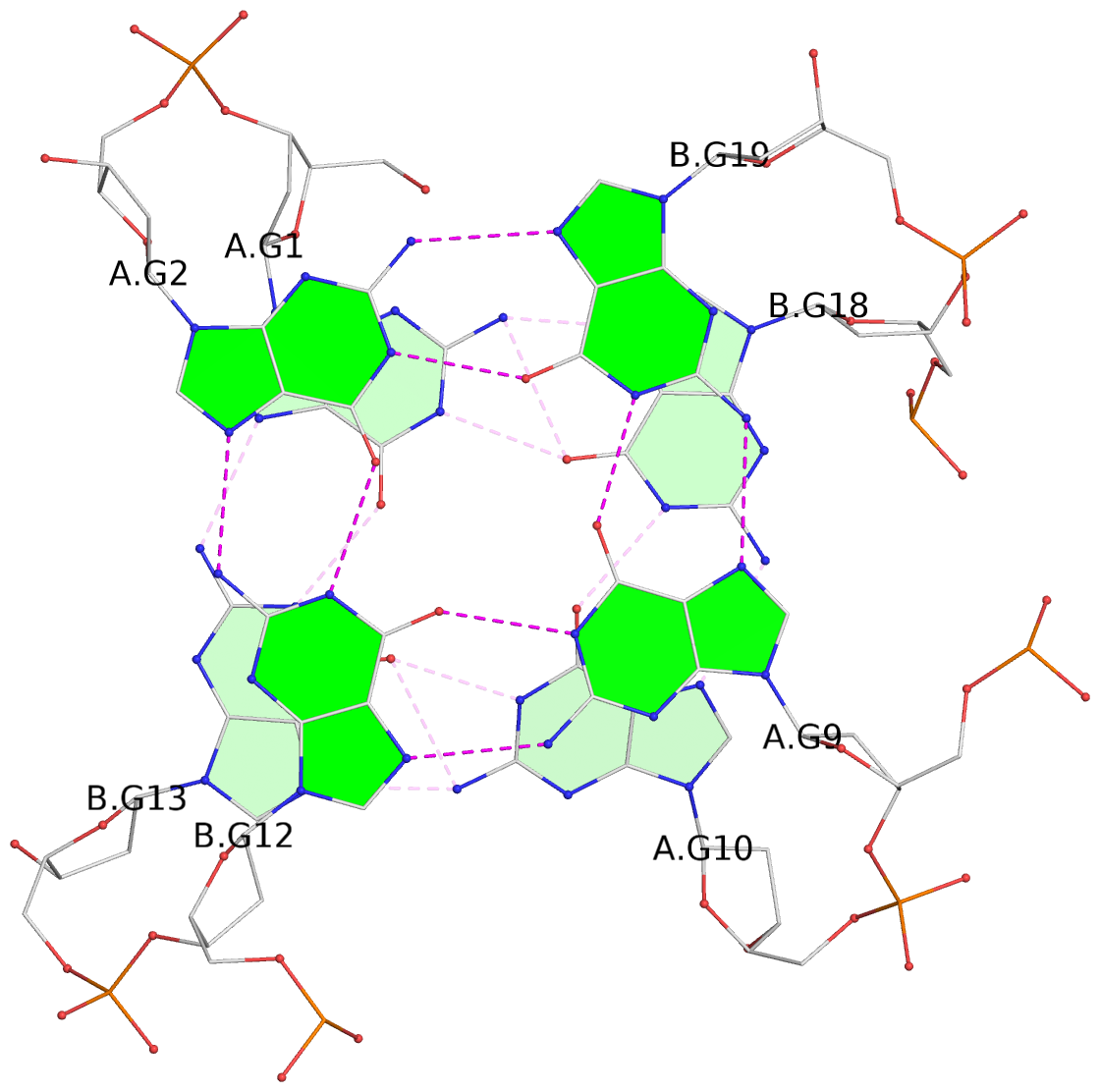
List of 3 G-tetrads
1 glyco-bond=s--s sugar=---- groove=w-n- planarity=0.409 type=bowl nts=4 GGGG A.DG1,B.DG13,A.DG10,B.DG18 2 glyco-bond=s--s sugar=.--- groove=w-n- planarity=0.158 type=planar nts=4 GGGG A.DG2,B.DG12,A.DG9,B.DG19 3 glyco-bond=-ss- sugar=---- groove=w-n- planarity=0.243 type=other nts=4 GGGG A.DG3,B.DG11,A.DG8,B.DG20
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, inter-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.