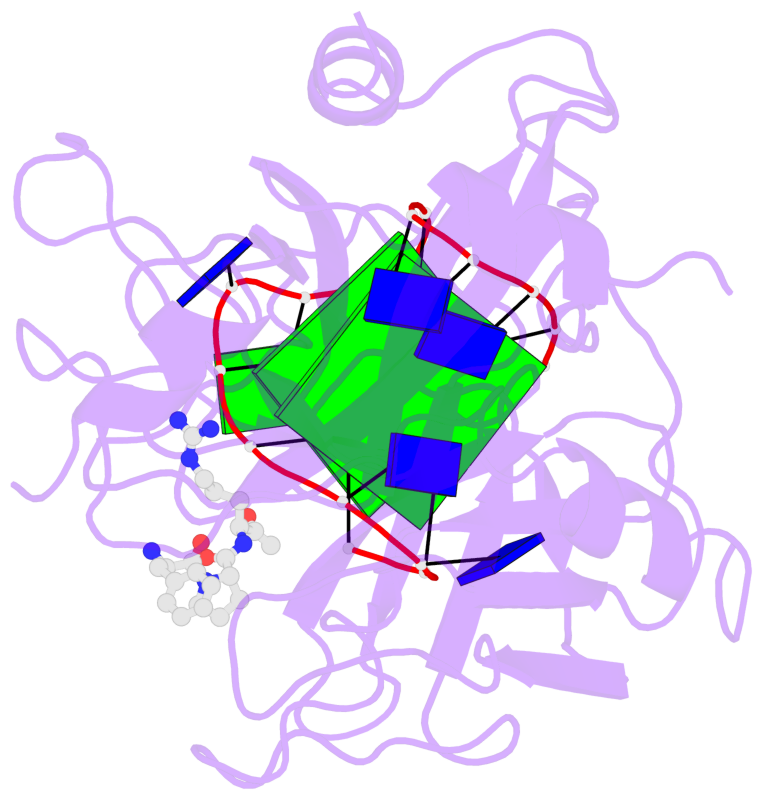
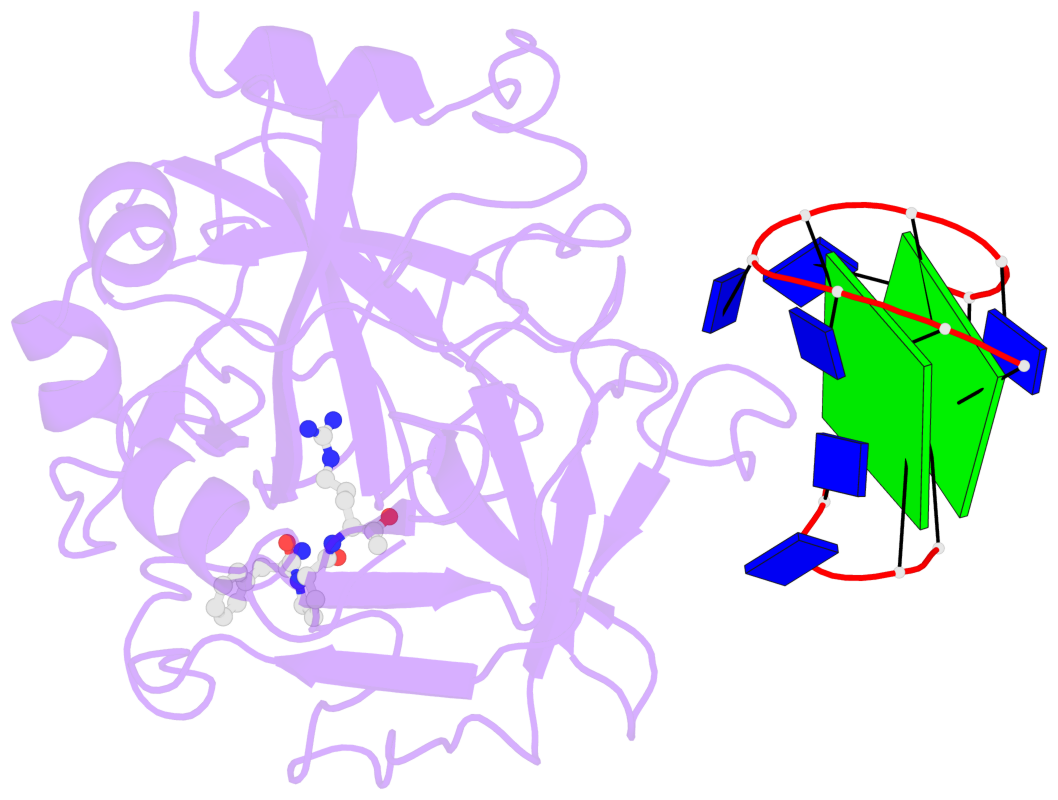
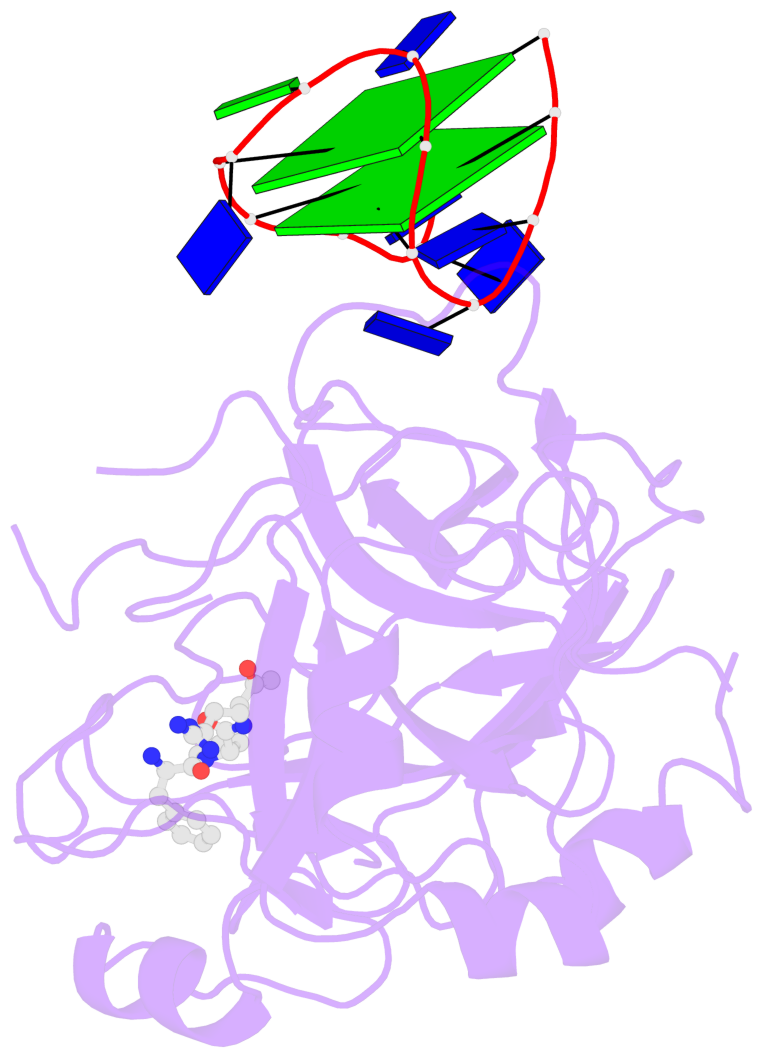
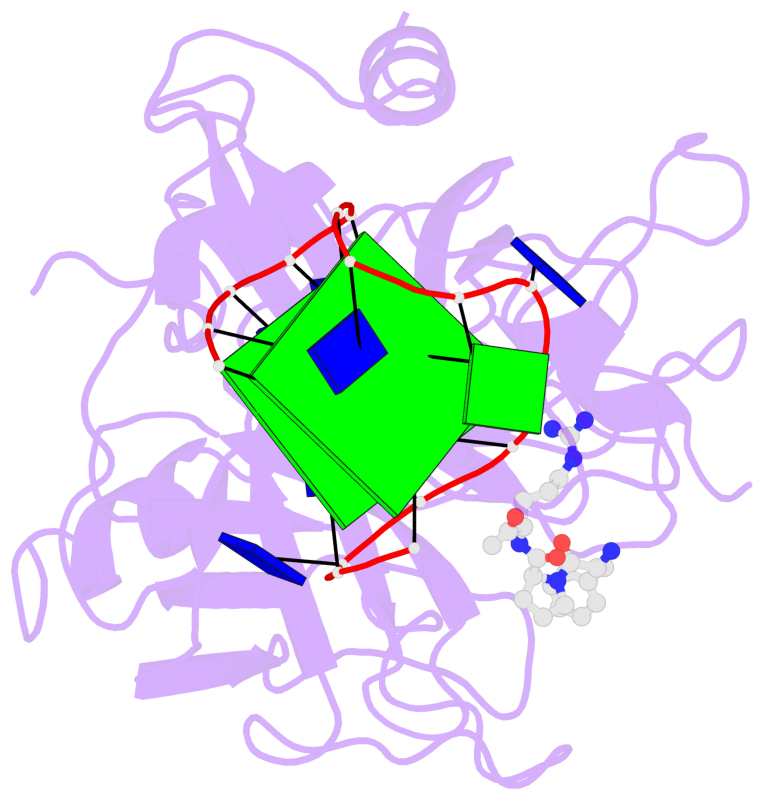
Detailed DSSR results for the G-quadruplex: PDB entry 1hao
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 1hao
- Class
- hydrolase-hydrolase inhibitor-DNA
- Method
- X-ray (2.8 Å)
- Summary
- Complex of human alpha-thrombin with a 15mer oligonucleotide ggttggtgtggttgg (based on NMR model of DNA)
- Reference
- Padmanabhan K, Tulinsky A (1996): "An ambiguous structure of a DNA 15-mer thrombin complex." Acta Crystallogr.,Sect.D, 52, 272-282. doi: 10.1107/S0907444995013977.
- Abstract
- The structure of a complex between thrombin and a GGTTGGTGTGGTTGG DNA 15-mer has been analyzed crystallographically. The solution NMR structure of the 15-mer has two stacked G-quartets similar to that found in the previous X-ray structure determination of the 15-mer-thrombin complex [Padmanabhan, Padmanabhan, Ferrara, Sadler & Tulinsky (1993). J. Biol. Chem. 268, 17651-17654]; the strand polarity, however, is reversed from that of the crystallographic structure. The structure of the complex here has been redetermined with better diffraction data confirming the previous crystallographic structure but also indicating that the NMR solution structure fits equally well. Both 15-mer complex structures refined to an R value of about 0.16 presenting a disconcerting ambiguity. Since the two 15-mer structures associate with thrombin in different ways (through the TGT loop in the X-ray and TT loop in the NMR model), other independent lines of physical or chemical evidence are required to resolve the ambiguity.
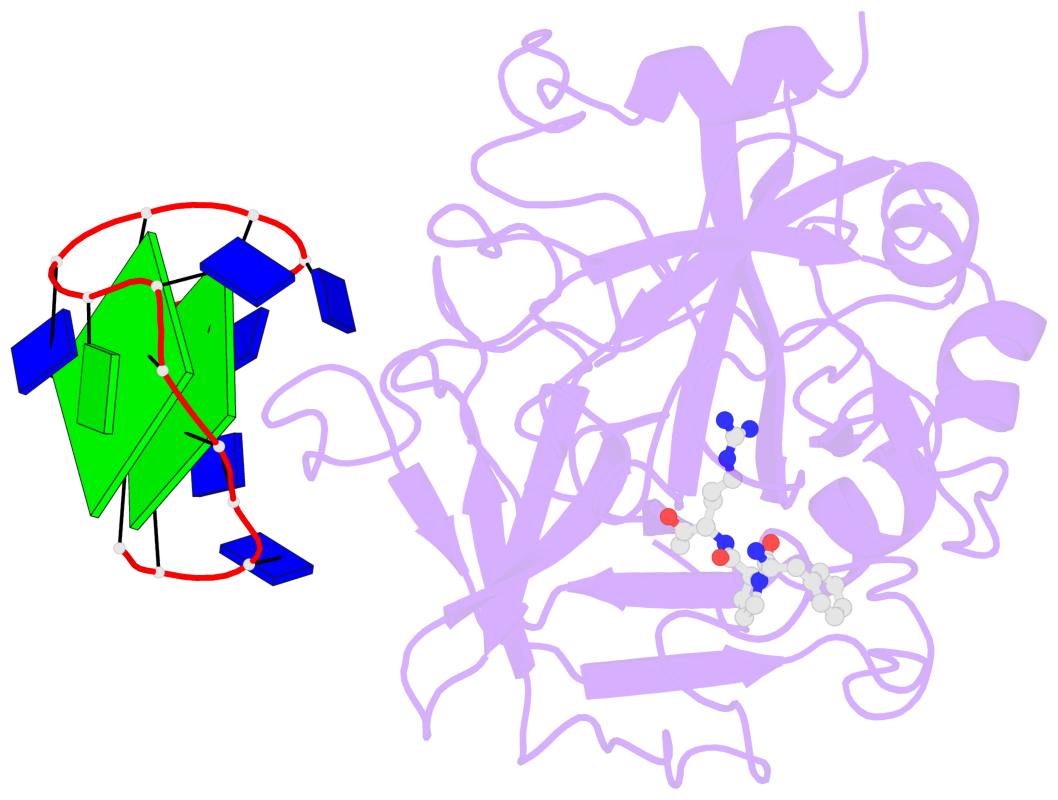
- G4 notes
- 2 G-tetrads, 1 G4 helix, 1 G4 stem, 2(+Ln+Lw+Ln), chair(2+2), UDUD
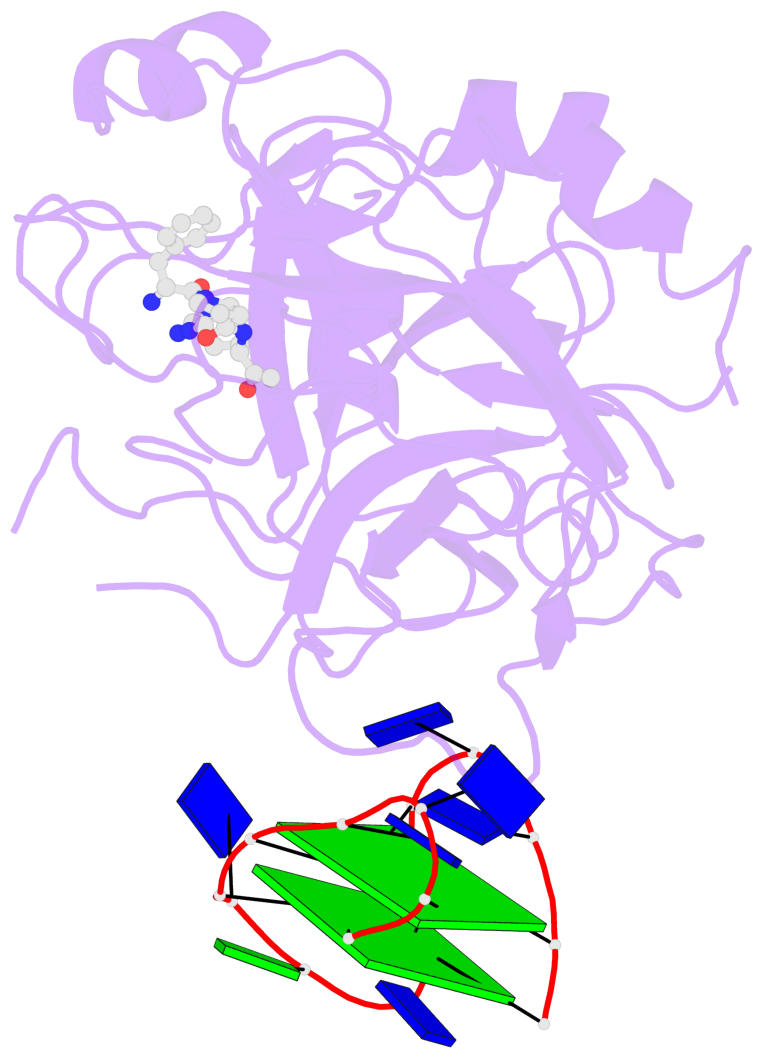
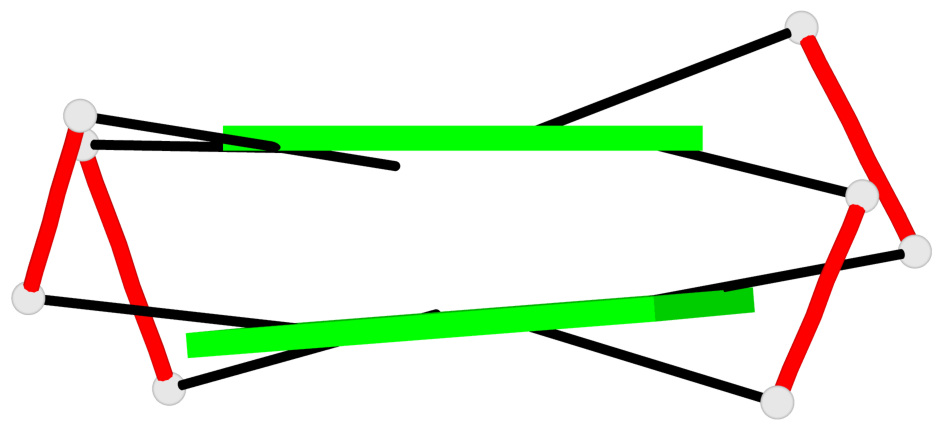
Base-block schematics in six views
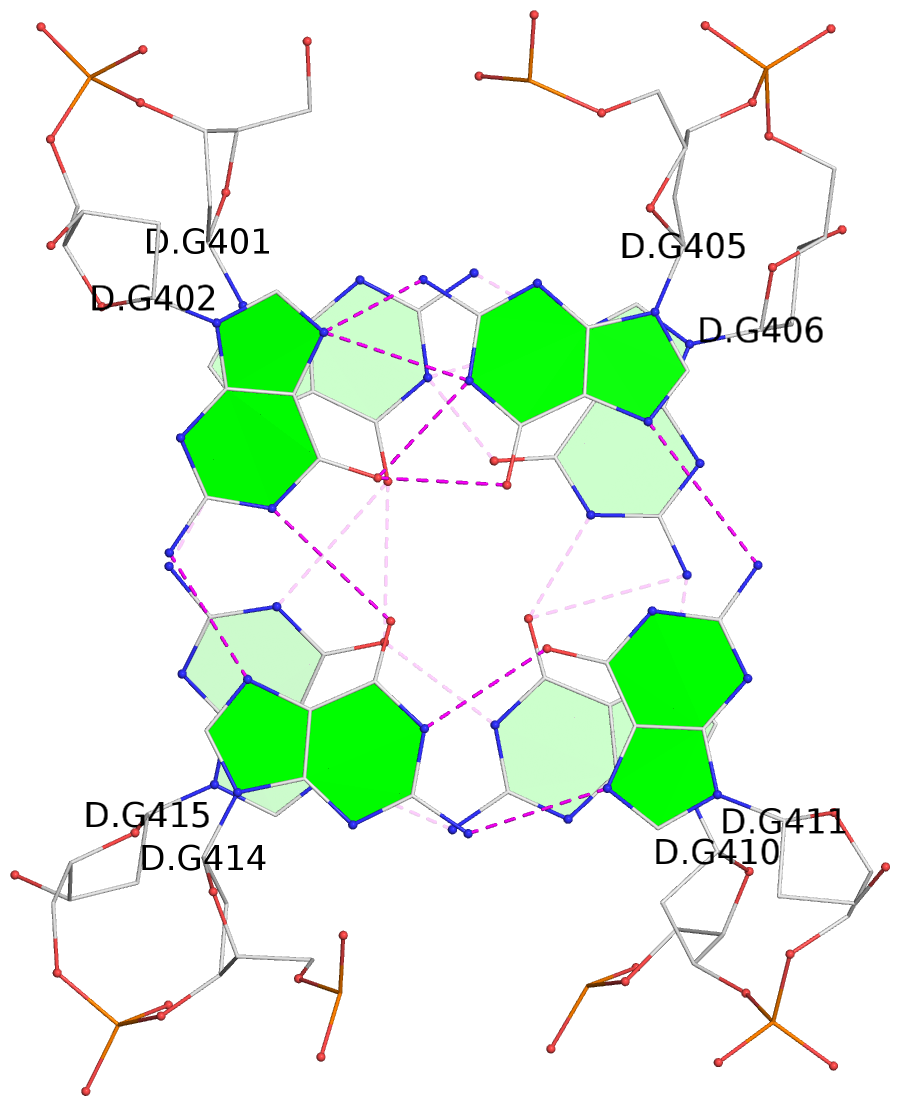
List of 2 G-tetrads
1 glyco-bond=s-s- sugar=---3 groove=wnwn planarity=0.373 type=other nts=4 GGGG D.DG401,D.DG415,D.DG410,D.DG406 2 glyco-bond=-s-s sugar=---. groove=wnwn planarity=0.441 type=other nts=4 GGGG D.DG402,D.DG414,D.DG411,D.DG405
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 2 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.