Detailed DSSR results for the G-quadruplex: PDB entry 1j6s
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 1j6s
- Class
- RNA
- Method
- X-ray (1.4 Å)
- Summary
- Crystal structure of an RNA tetraplex (ugaggu)4 with a-tetrads, g-tetrads, u-tetrads and g-u octads
- Reference
- Pan B, Xiong Y, Shi K, Deng J, Sundaralingam M (2003): "Crystal structure of an RNA purine-rich tetraplex containing adenine tetrads: implications for specific binding in RNA tetraplexes." Structure, 11, 815-823. doi: 10.1016/S0969-2126(03)00107-2.
- Abstract
- Purine-rich regions in DNA and RNA may contain both guanines and adenines, which have various biological functions. Here we report the crystal structure of an RNA purine-rich fragment containing both guanine and adenine at 1.4 A resolution. Adenines form an adenine tetrad in the N6-H em leader N7 conformation. Substitution of an adenine tetrad in the guanine tetraplex does not change the global conformation but introduces irregularity in both the hydrogen bonding interaction pattern in the groove and the metal ion binding pattern in the central cavity of the tetraplex. The irregularity in groove binding may be critical for specific binding in tetraplexes. The formation of G-U octads provides a mechanism for interaction in the groove. Ba(2+) ions prefer to bind guanine tetrads, and adenine tetrads can only be bound by Na(+) ions, illustrating the binding selectivity of metal ions for the tetraplex.
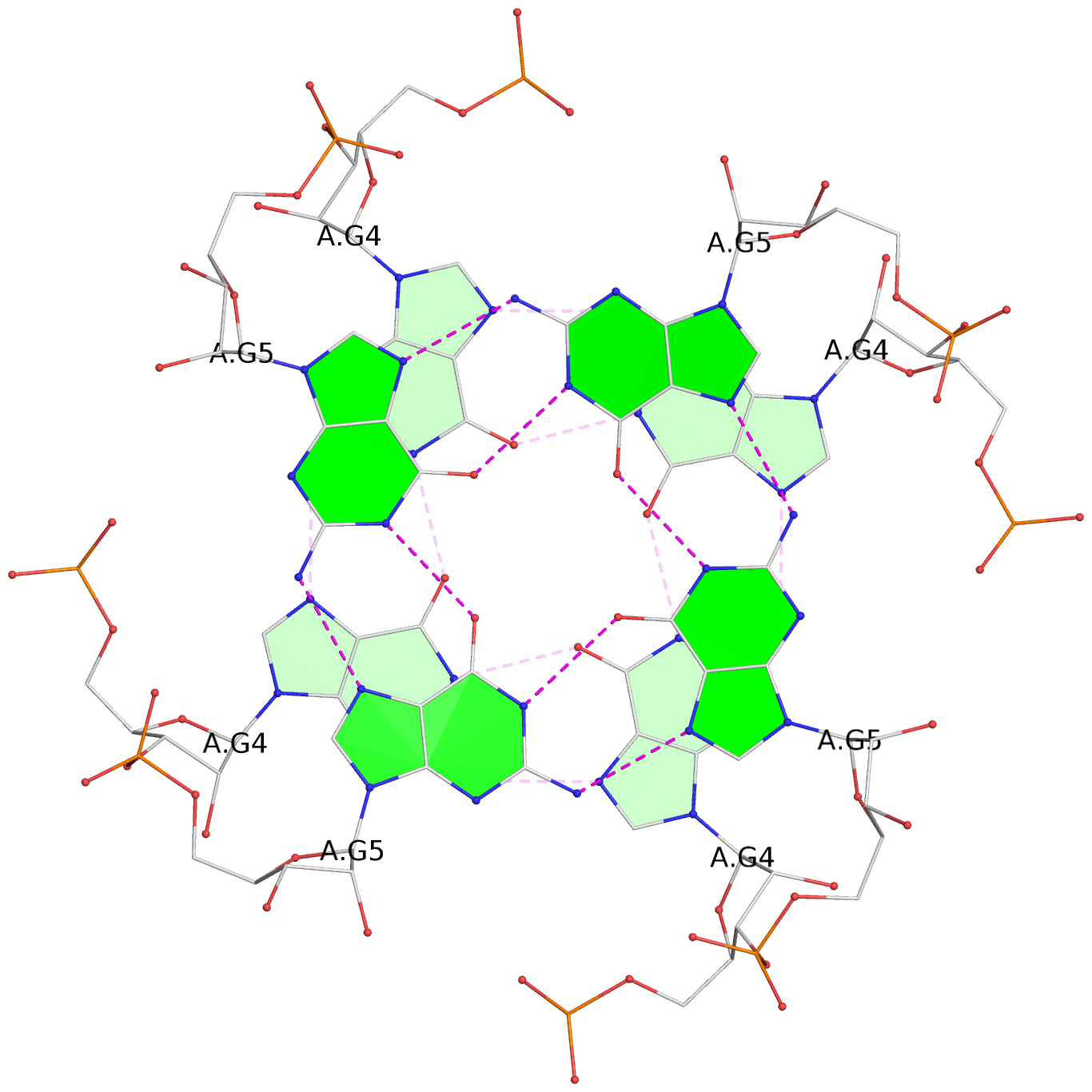
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, parallel(4+0), UUUU
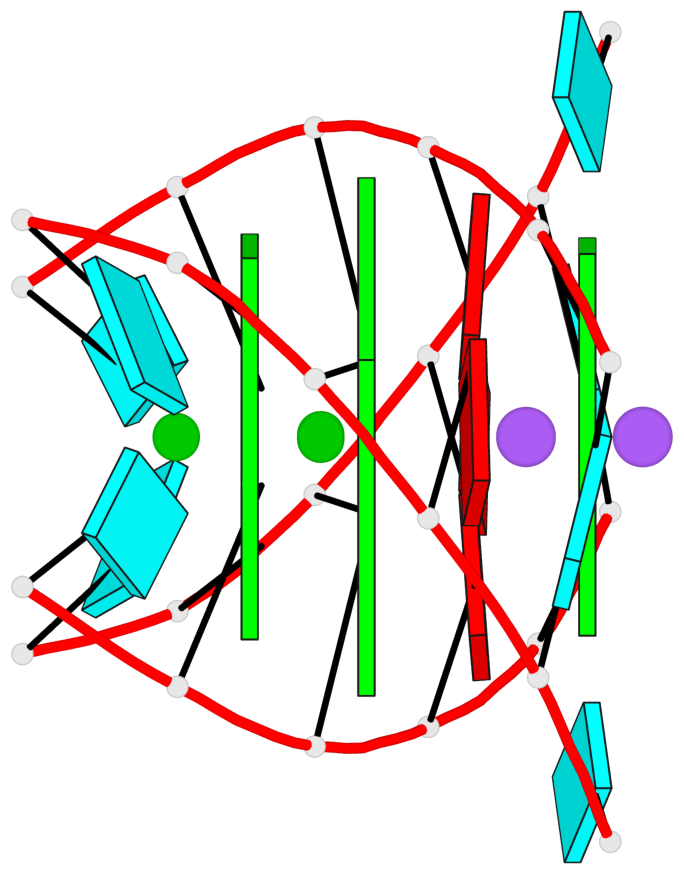
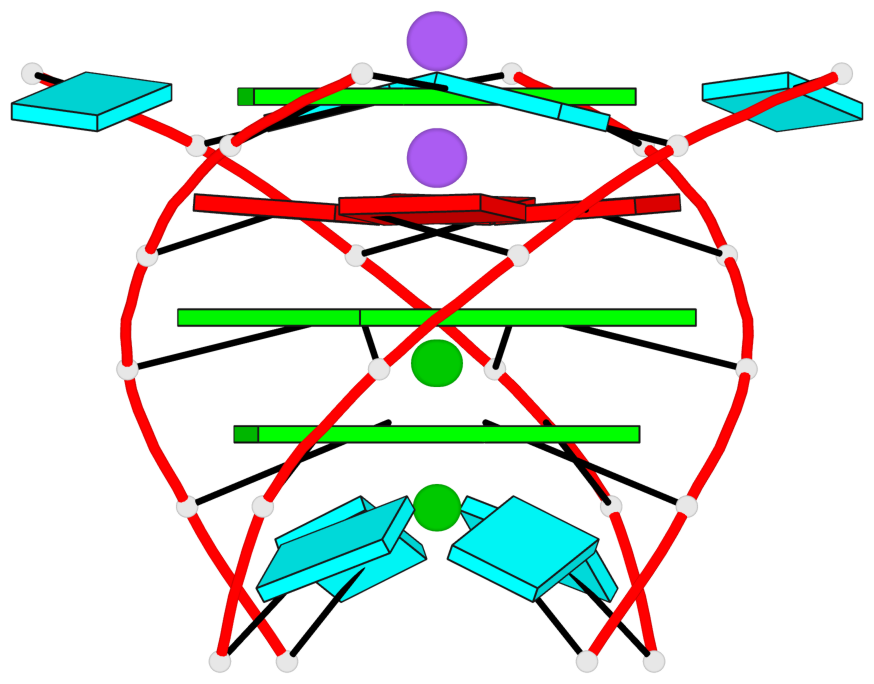
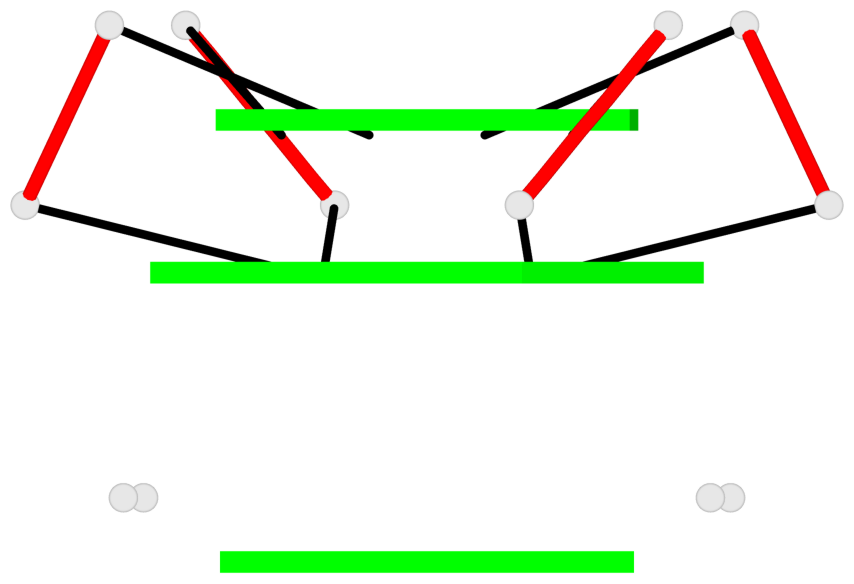
Base-block schematics in six views
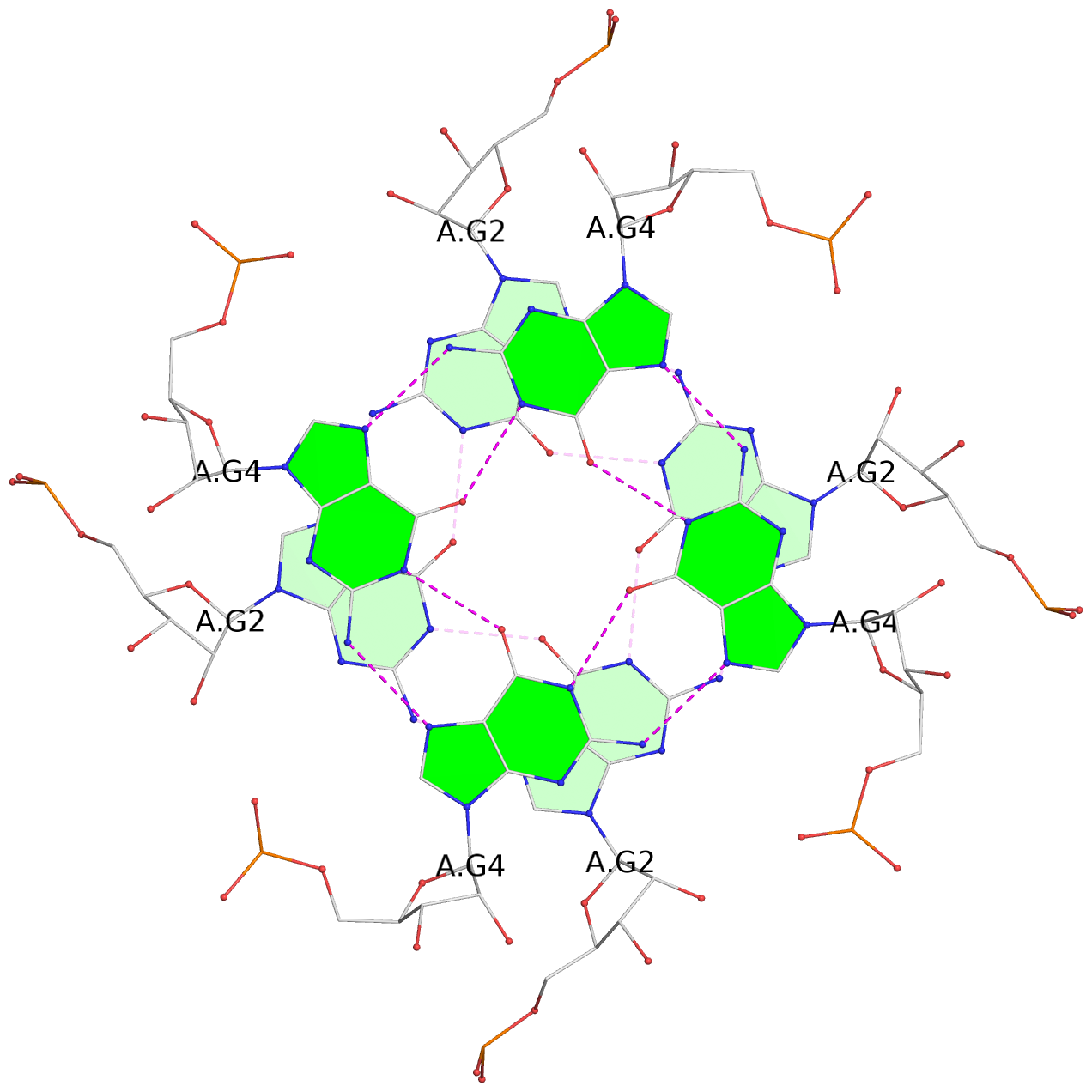
List of 3 G-tetrads
1 glyco-bond=---- sugar=3333 groove=---- planarity=0.101 type=planar nts=4 GGGG 1:A.G2,4:A.G2,2:A.G2,3:A.G2 2 glyco-bond=---- sugar=3333 groove=---- planarity=0.067 type=planar nts=4 GGGG 1:A.G4,4:A.G4,2:A.G4,3:A.G4 3 glyco-bond=---- sugar=3333 groove=---- planarity=0.285 type=bowl nts=4 GGGG 1:A.G5,4:A.G5,2:A.G5,3:A.G5
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, inter-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.