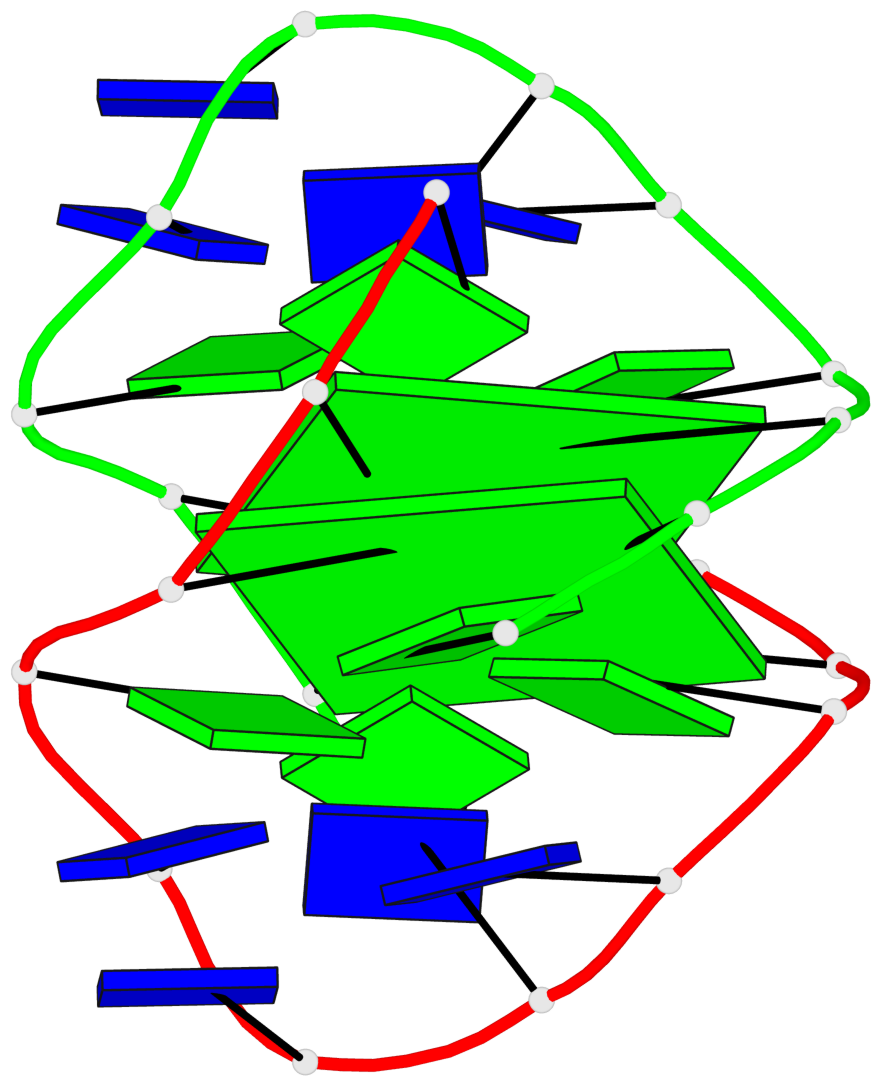
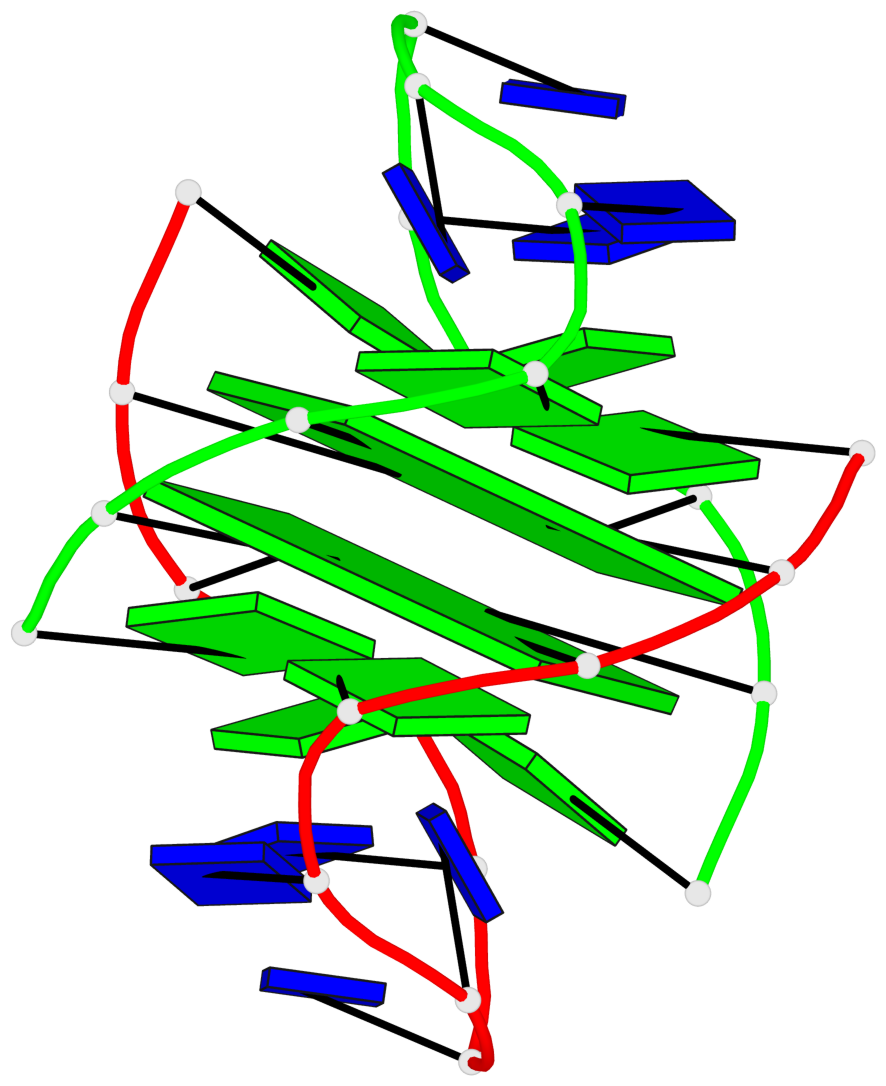
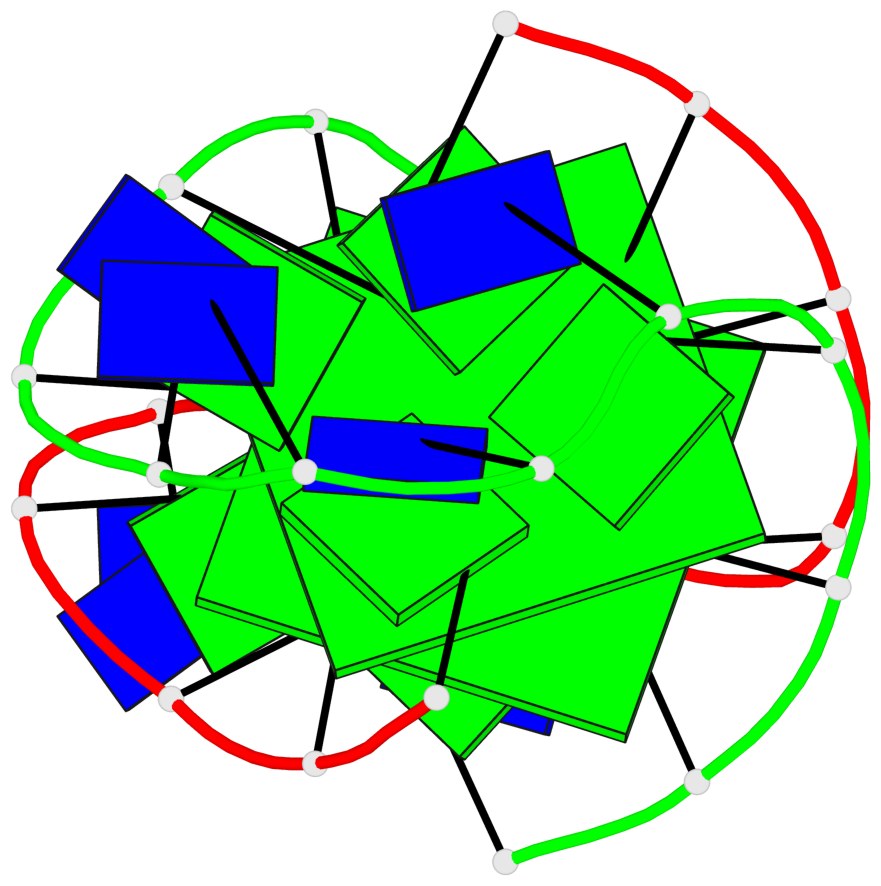
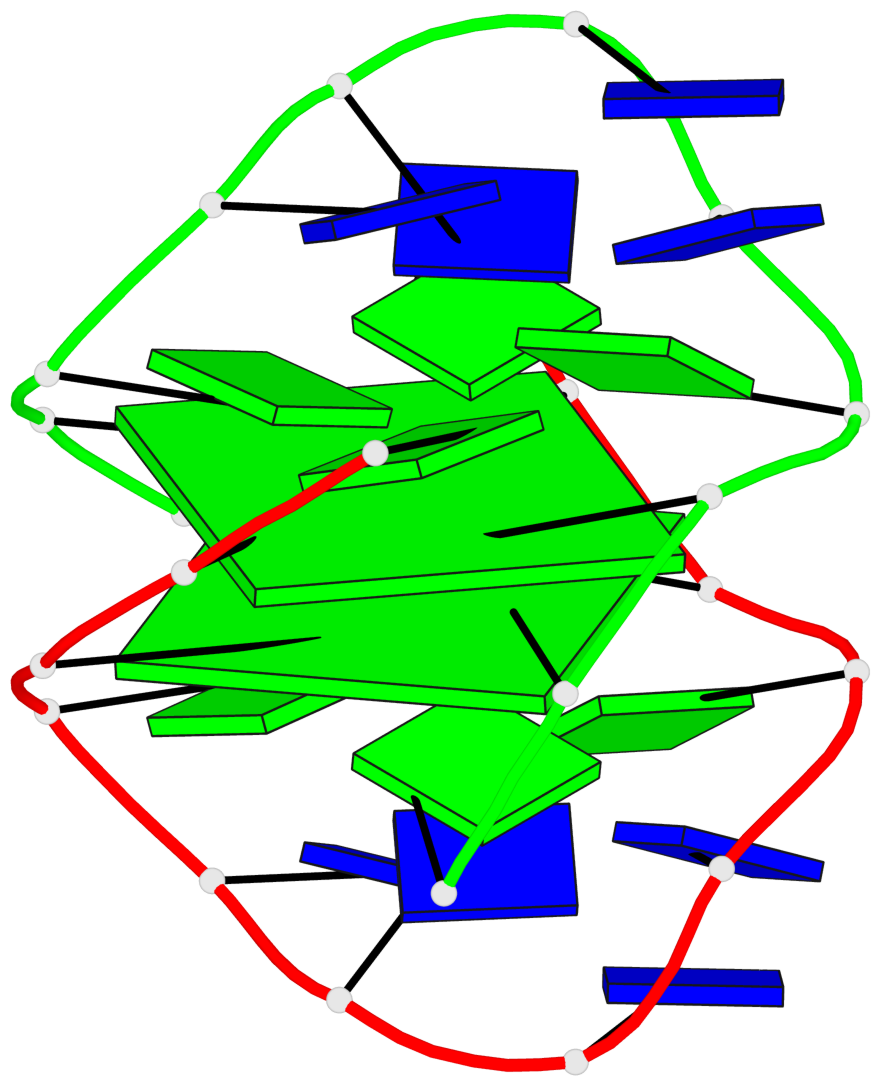
Detailed DSSR results for the G-quadruplex: PDB entry 1k4x
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 1k4x
- Class
- DNA
- Method
- NMR
- Summary
- Potassium form of oxy-1.5 quadruplex DNA
- Reference
- Schultze P, Hud NV, Smith FW, Feigon J (1999): "The effect of sodium, potassium and ammonium ions on the conformation of the dimeric quadruplex formed by the Oxytricha nova telomere repeat oligonucleotide d(G(4)T(4)G(4))." Nucleic Acids Res., 27, 3018-3028. doi: 10.1093/nar/27.15.3018.
- Abstract
- The DNA sequence d(G(4)T(4)G(4)) [Oxy-1.5] consists of 1.5 units of the repeat in telomeres of Oxytricha nova and has been shown by NMR and X-ray crystallographic analysis to form a dimeric quadruplex structure with four guanine-quartets. However, the structure reported in the X-ray study has a fundamentally different conformation and folding topology compared to the solution structure. In order to elucidate the possible role of different counterions in this discrepancy and to investigate the conformational effects and dynamics of ion binding to G-quadruplex DNA, we compare results from further experiments using a variety of counterions, namely K(+), Na(+)and NH(4)(+). A detailed structure determination of Oxy-1.5 in solution in the presence of K(+)shows the same folding topology as previously reported with the same molecule in the presence of Na(+). Both conformations are symmetric dimeric quadruplexes with T(4)loops which span the diagonal of the end quartets. The stack of quartets shows only small differences in the presence of K(+)versus Na(+)counterions, but the T(4)loops adopt notably distinguishable conformations. Dynamic NMR analysis of the spectra of Oxy-1.5 in mixed Na(+)/K(+)solution reveals that there are at least three K(+)binding sites. Additional experiments in the presence of NH(4)(+)reveal the same topology and loop conformation as in the K(+)form and allow the direct localization of three central ions in the stack of quartets and further show that there are no specific NH(4)(+)binding sites in the T(4)loop. The location of bound NH(4)(+)with respect to the expected coordination sites for Na(+)binding provides a rationale for the difference observed for the structure of the T(4)loop in the Na(+)form, with respect to that observed for the K(+)and NH(4)(+)forms.
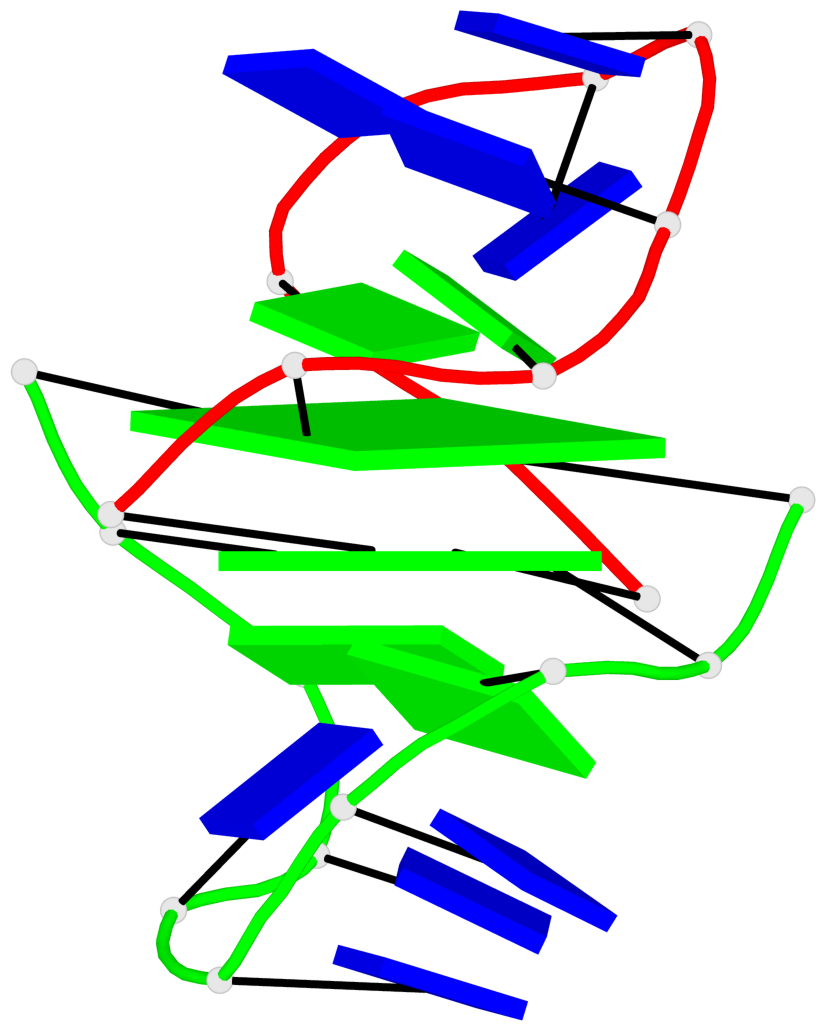
- G4 notes
- 2 G-tetrads, 1 G4 helix, 1 G4 stem, (2+2), UDDU
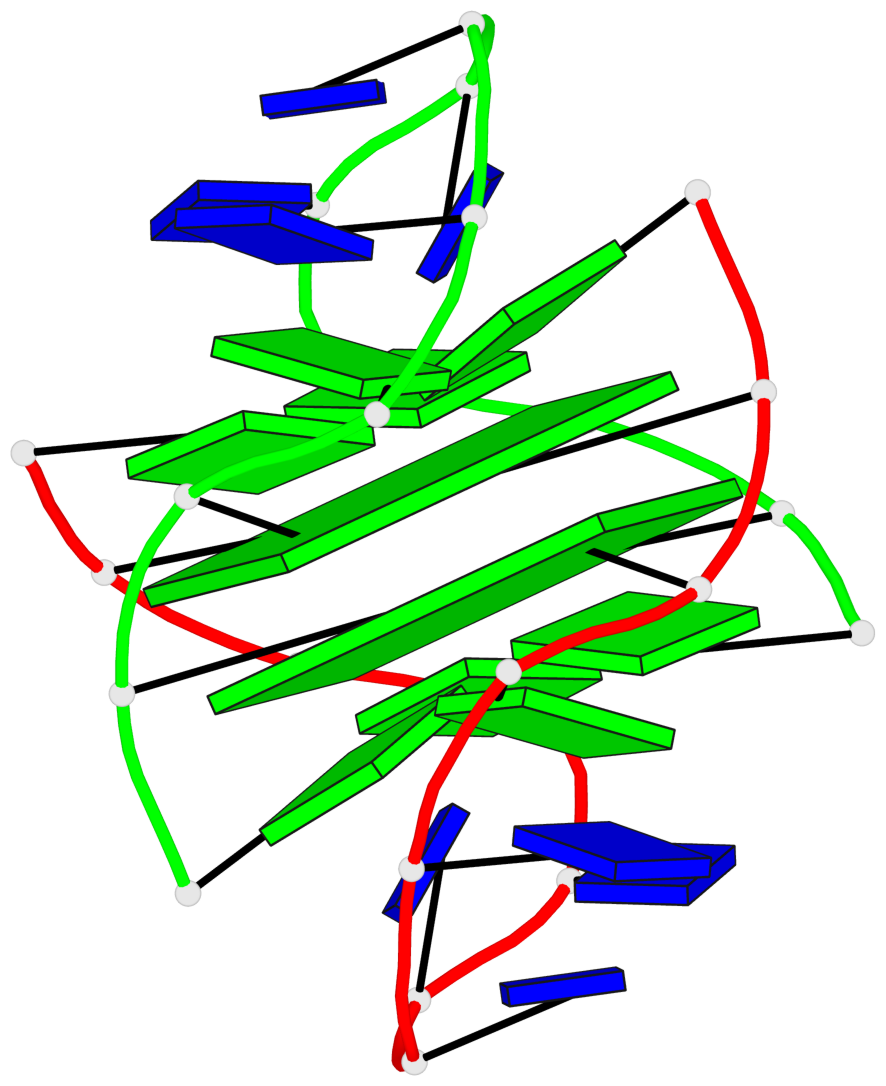
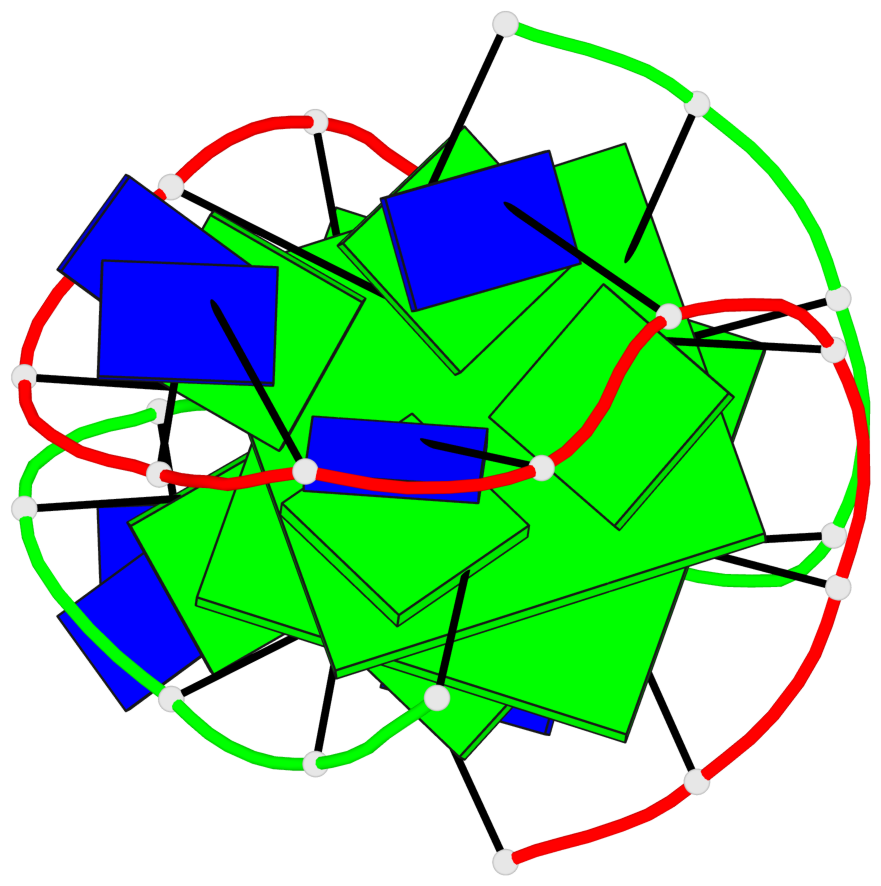
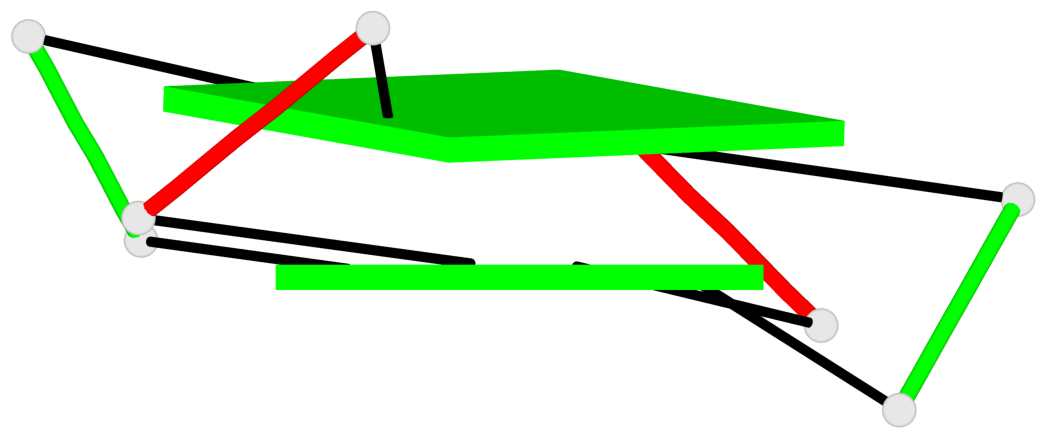
Base-block schematics in six views
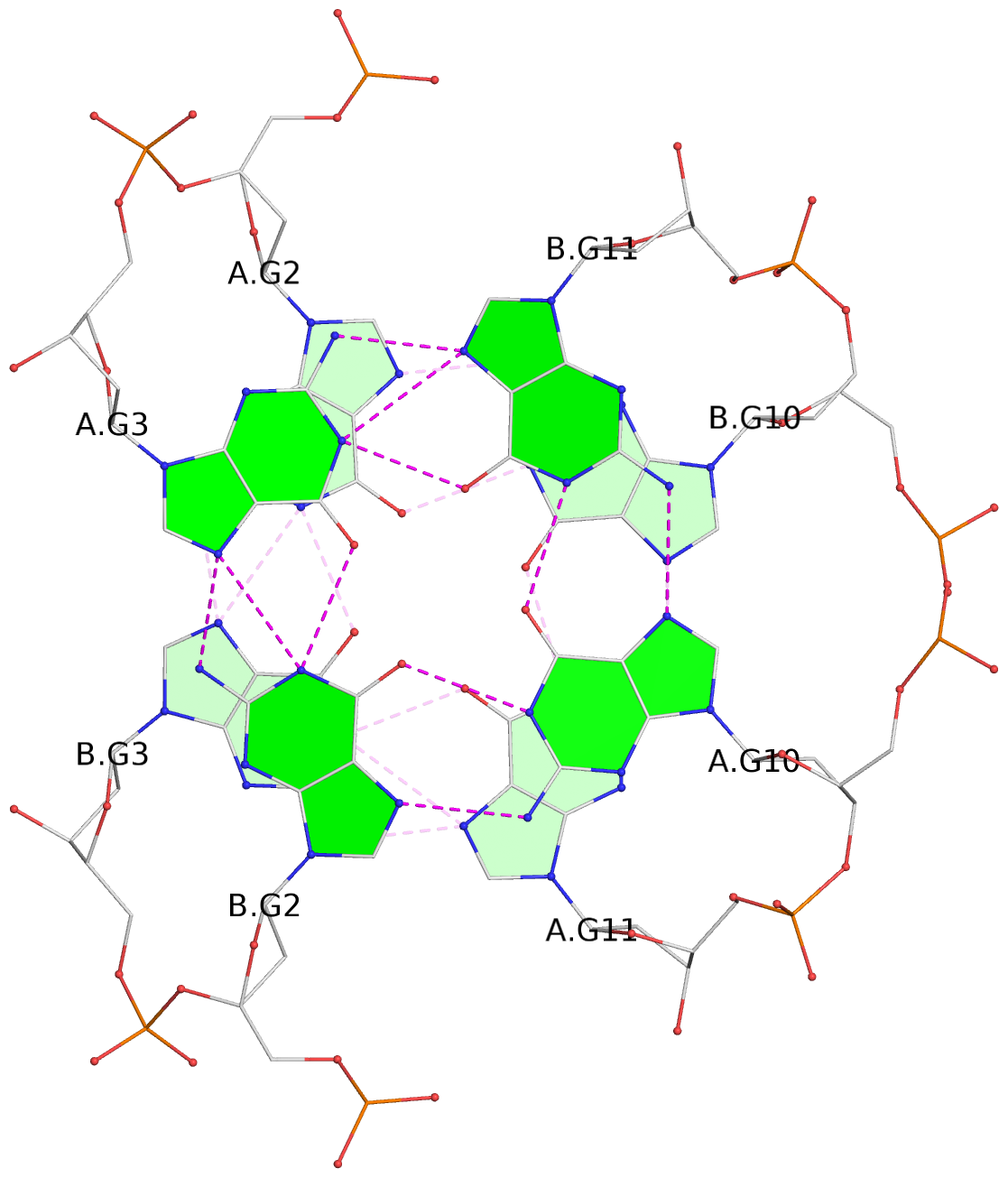
List of 2 G-tetrads
1 glyco-bond=-ss- sugar=---- groove=w-n- planarity=0.345 type=other nts=4 GGGG A.DG2,B.DG3,A.DG11,B.DG10 2 glyco-bond=s--s sugar=---- groove=w-n- planarity=0.345 type=other nts=4 GGGG A.DG3,B.DG2,A.DG10,B.DG11
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 2 G-tetrad layers, inter-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.