Detailed DSSR results for the G-quadruplex: PDB entry 1l1h
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 1l1h
- Class
- DNA
- Method
- X-ray (1.75 Å)
- Summary
- Crystal structure of the quadruplex DNA-drug complex
- Reference
- Haider SM, Parkinson GN, Neidle S (2003): "Structure of a G-quadruplex-Ligand Complex." J.Mol.Biol., 326, 117-125. doi: 10.1016/S0022-2836(02)01354-2.
- Abstract
- Stabilisation of G-quadruplex structures formed from telomeric DNA, by means of quadruplex-selective ligands, is a means of inhibiting the telomerase enzyme from catalysing the synthesis of telomeric DNA repeats. In order to understand the molecular basis of ligand-quadruplex recognition, the crystal structure has been determined of such a complex, at 1.75A resolution. This complex is between a dimeric antiparallel G-quadruplex formed from the Oxytricha nova telomeric DNA sequence d(GGGGTTTTGGGG), and a di-substituted aminoalkylamido acridine compound. The structure shows that the acridine moiety is bound at one end of the stack of G-quartets, within one of the thymine loops. It is held in place by a combination of stacking interactions and specific hydrogen bonds with thymine bases. The stability of the ligand in this binding site has been confirmed by a 2ns molecular dynamics simulation.
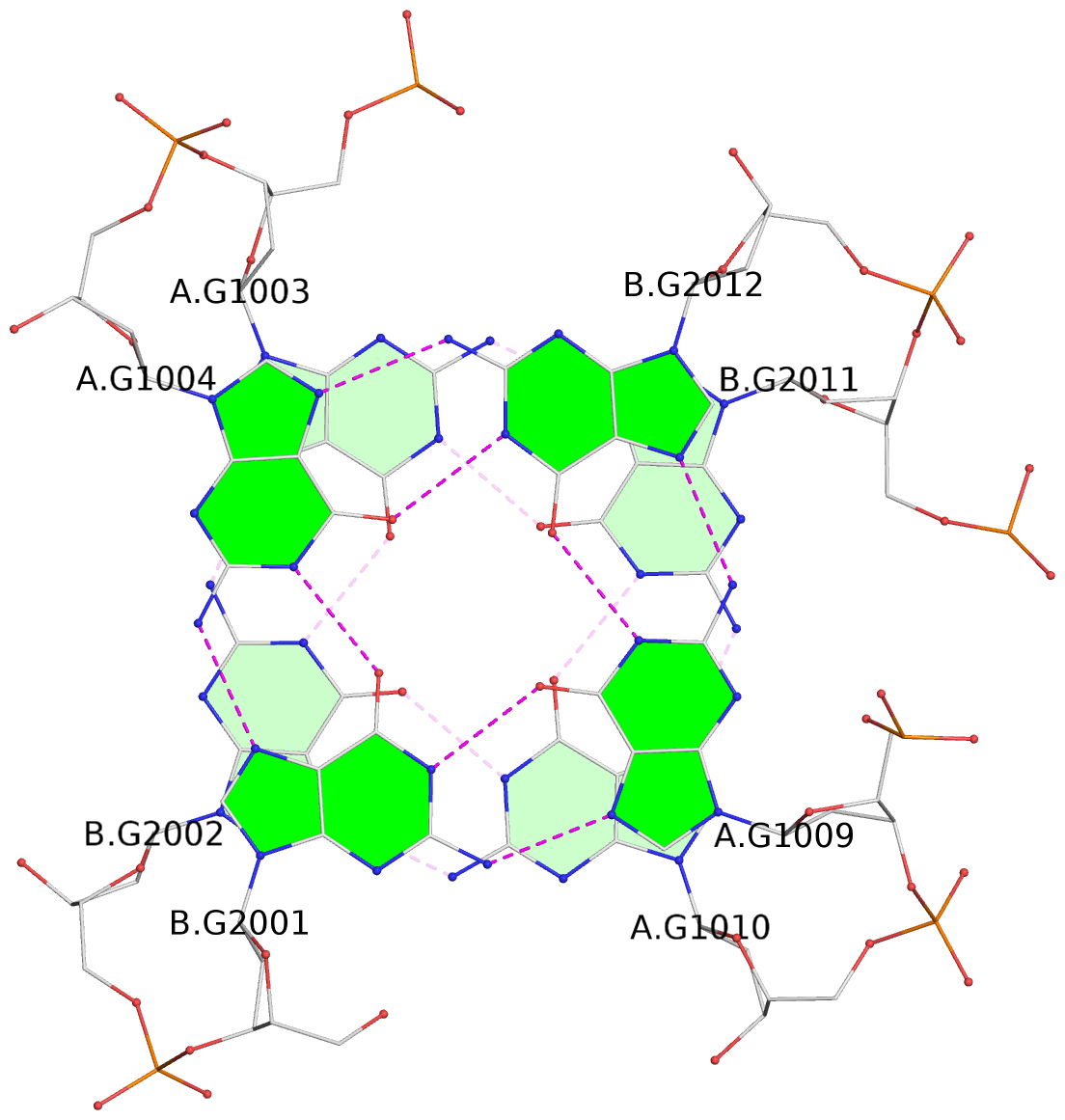
- G4 notes
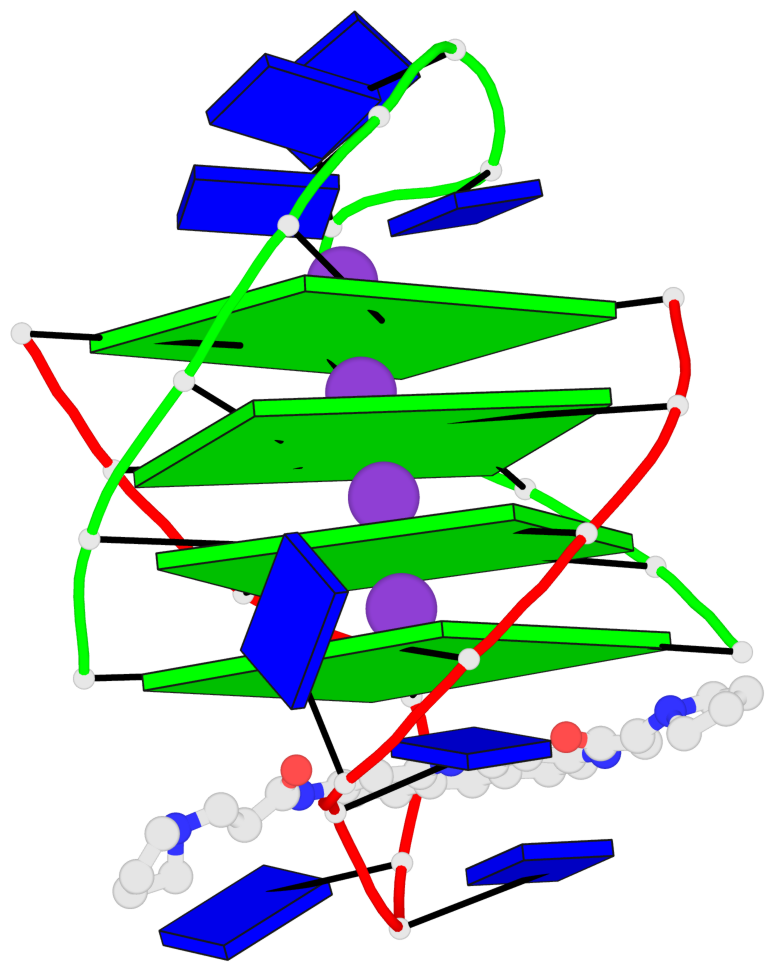
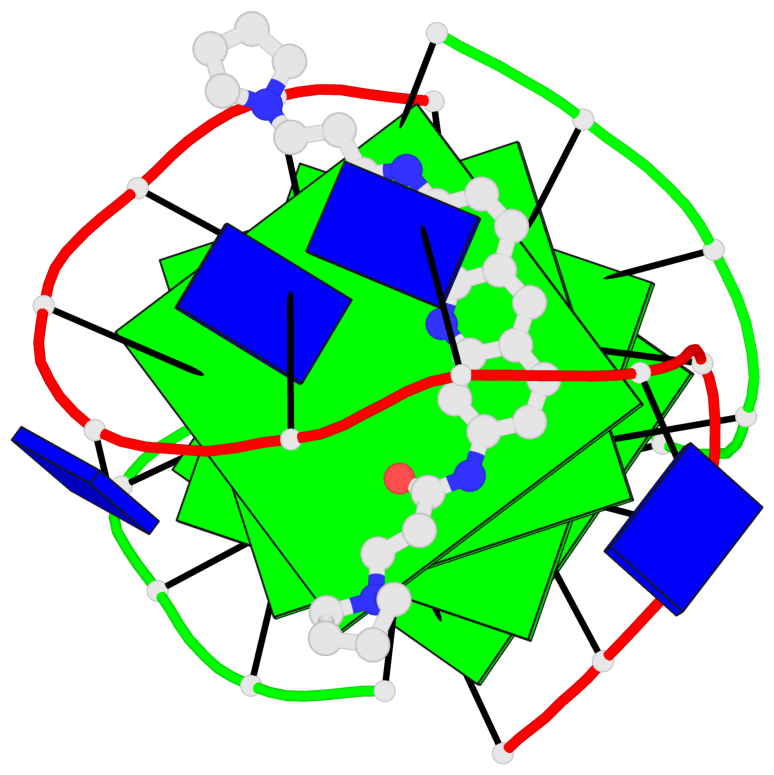
- 4 G-tetrads, 1 G4 helix, 1 G4 stem, (2+2), UDDU
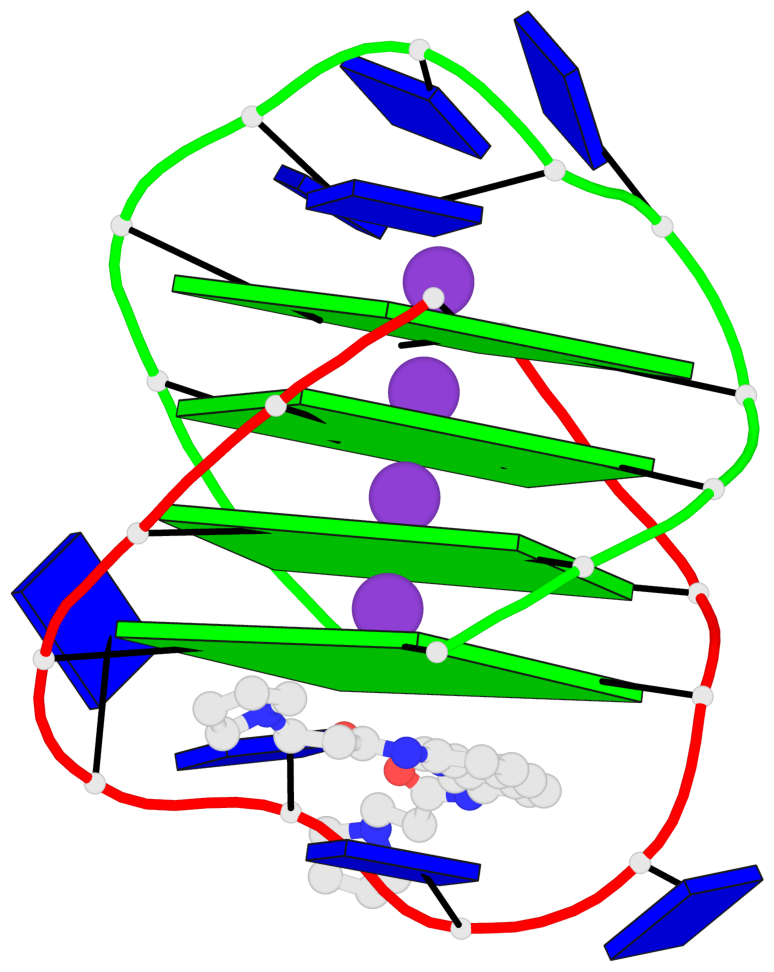
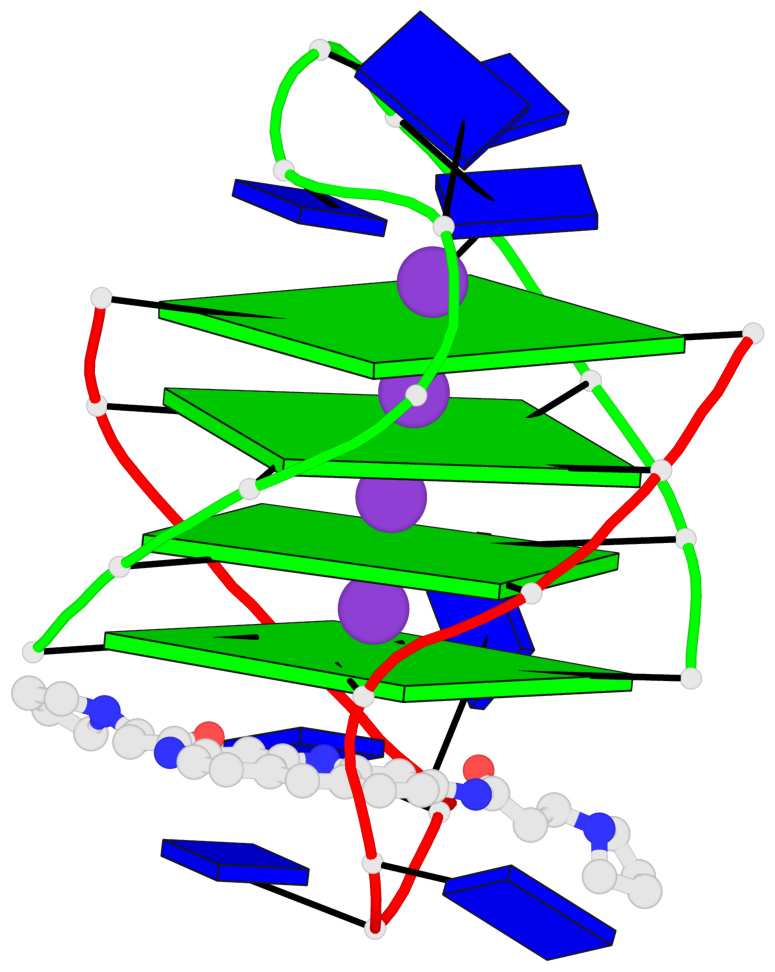
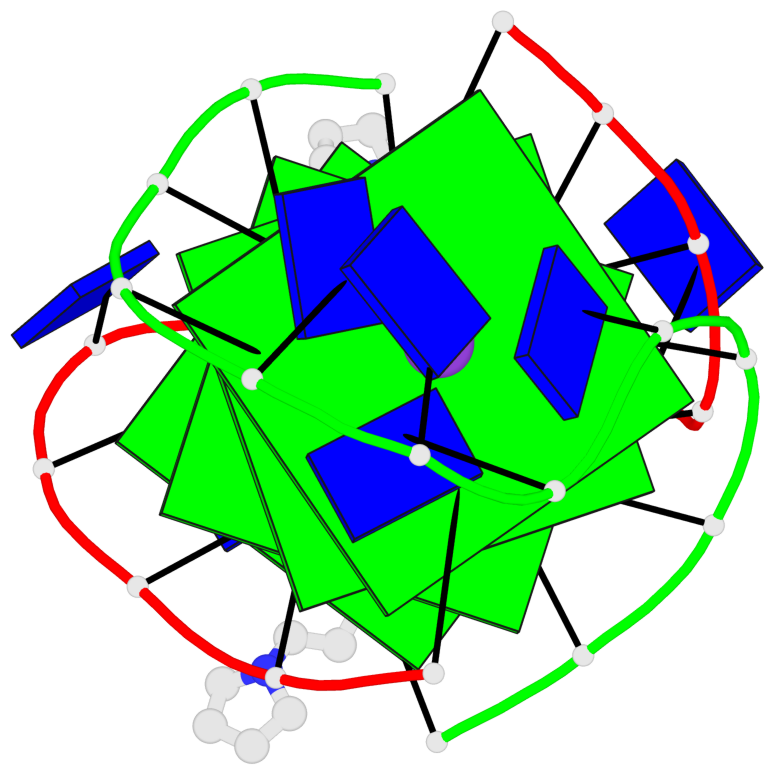
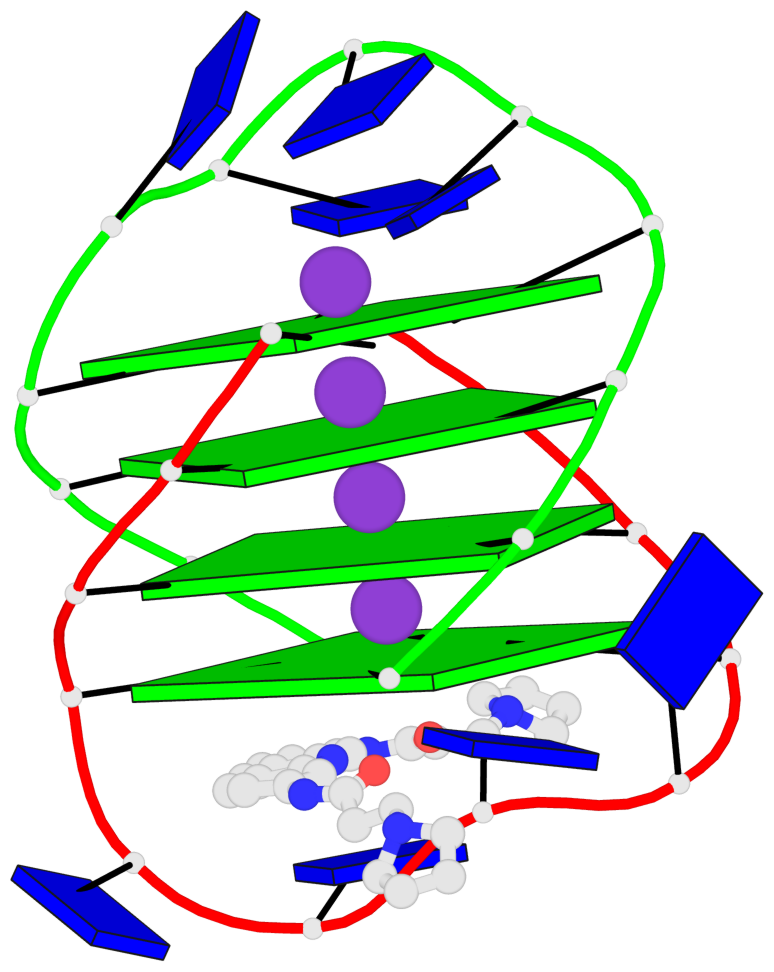
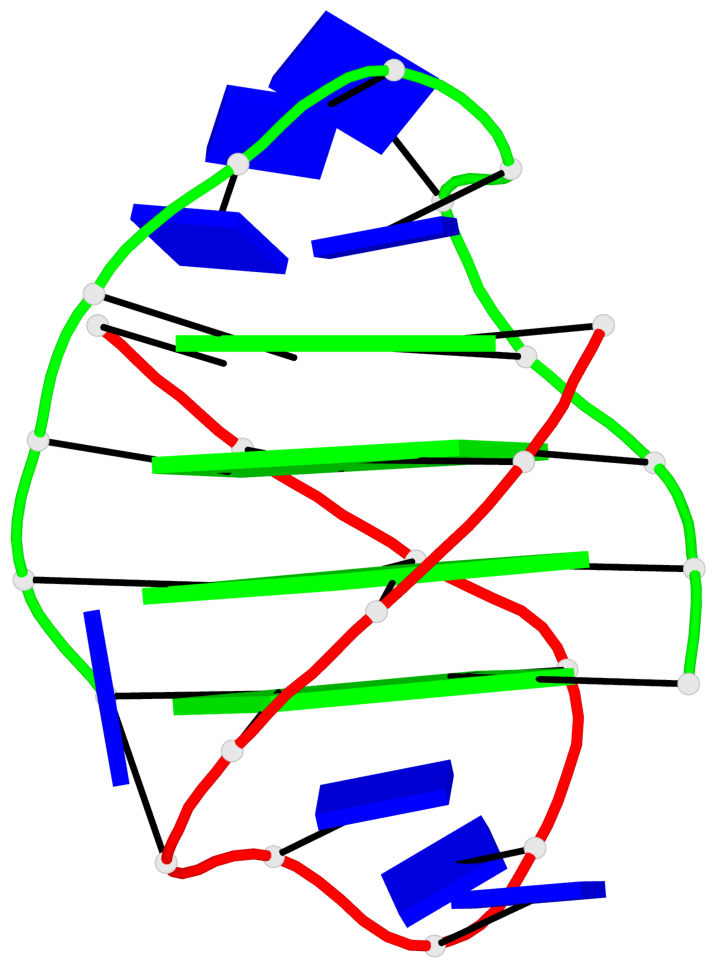
Base-block schematics in six views
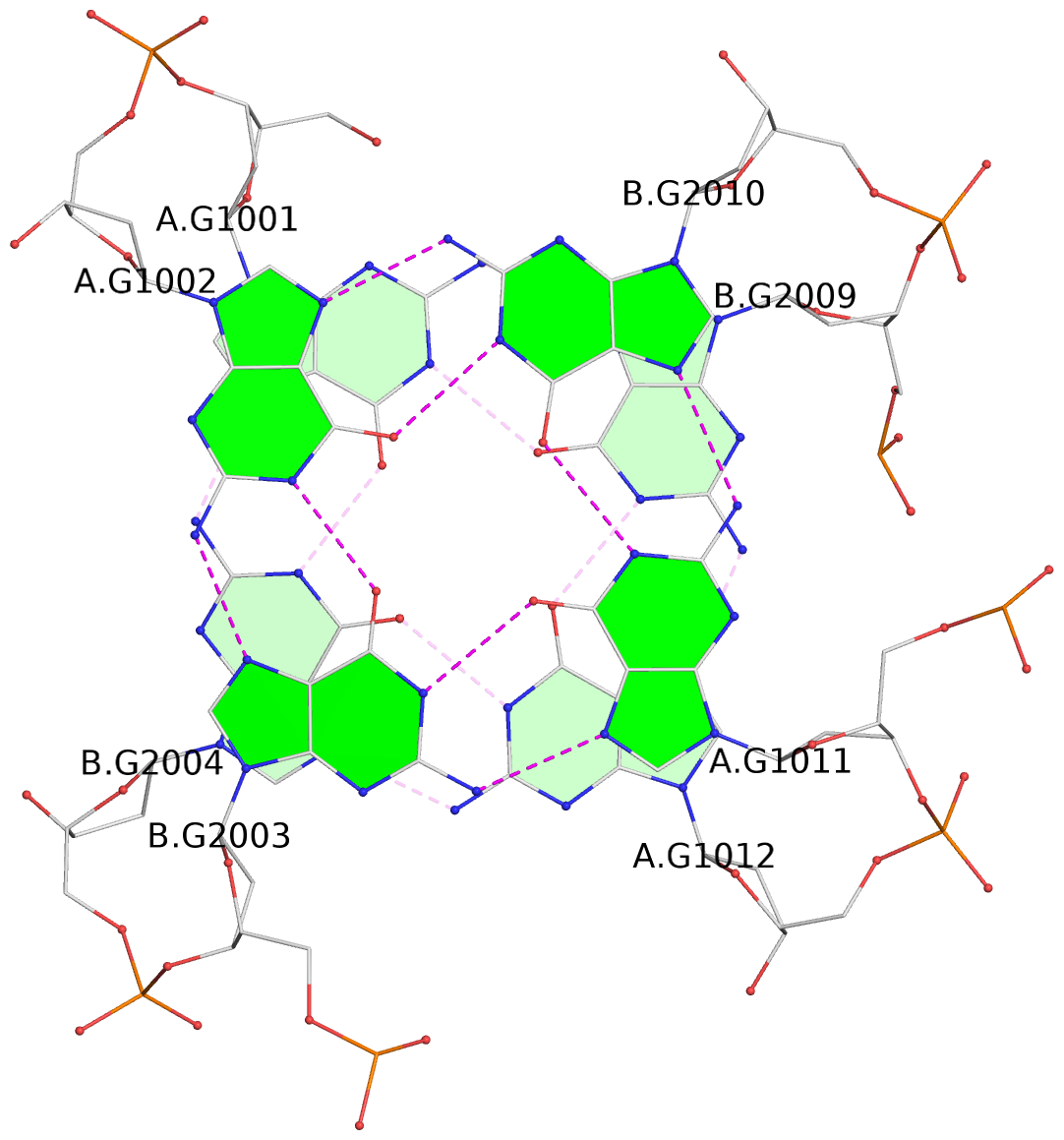
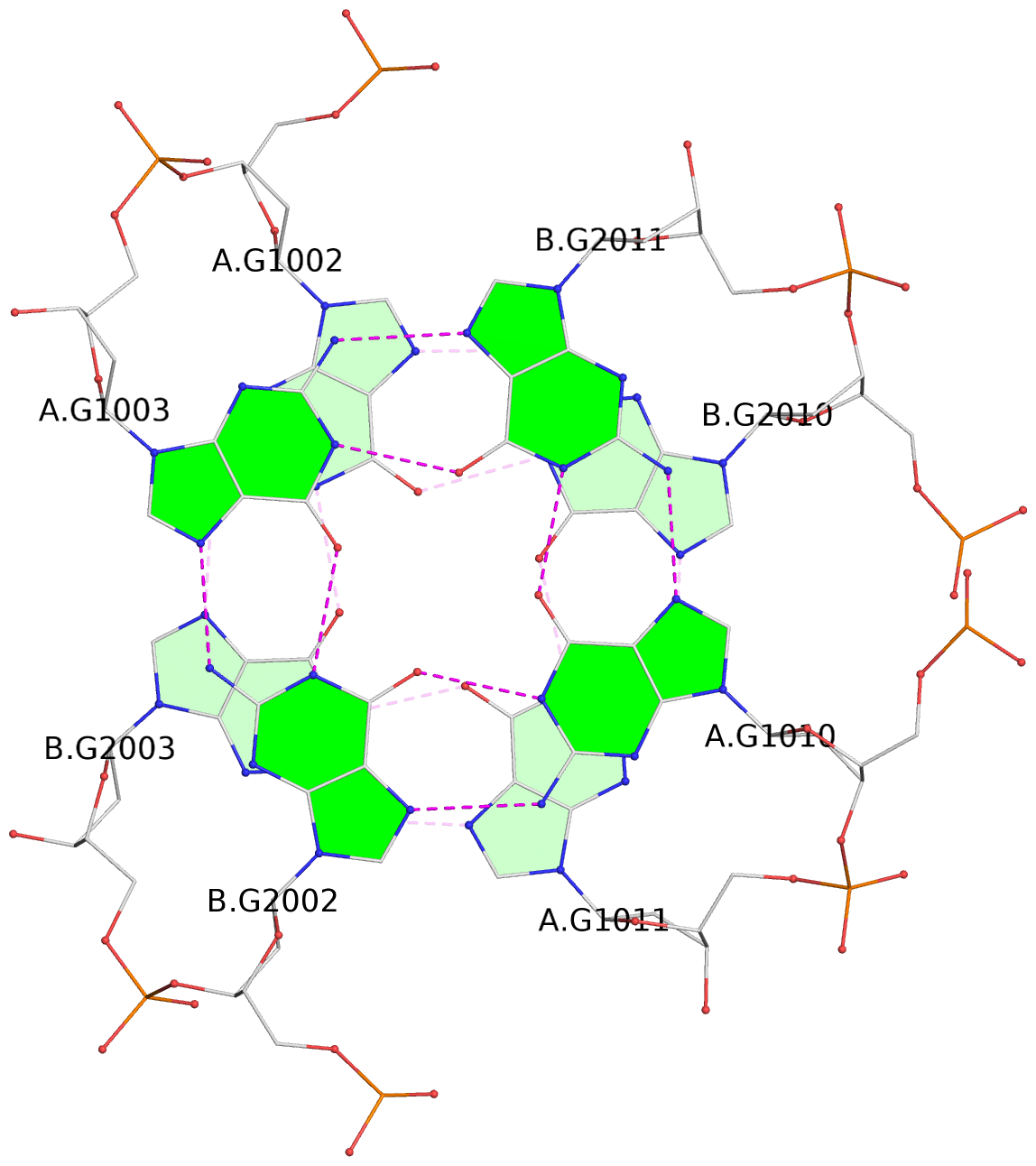
List of 4 G-tetrads
1 glyco-bond=s--s sugar=---- groove=w-n- planarity=0.279 type=bowl nts=4 GGGG A.DG1001,B.DG2004,A.DG1012,B.DG2009 2 glyco-bond=-ss- sugar=---- groove=w-n- planarity=0.192 type=other nts=4 GGGG A.DG1002,B.DG2003,A.DG1011,B.DG2010 3 glyco-bond=s--s sugar=---- groove=w-n- planarity=0.171 type=other nts=4 GGGG A.DG1003,B.DG2002,A.DG1010,B.DG2011 4 glyco-bond=-ss- sugar=---- groove=w-n- planarity=0.202 type=other nts=4 GGGG A.DG1004,B.DG2001,A.DG1009,B.DG2012
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 4 G-tetrad layers, inter-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.