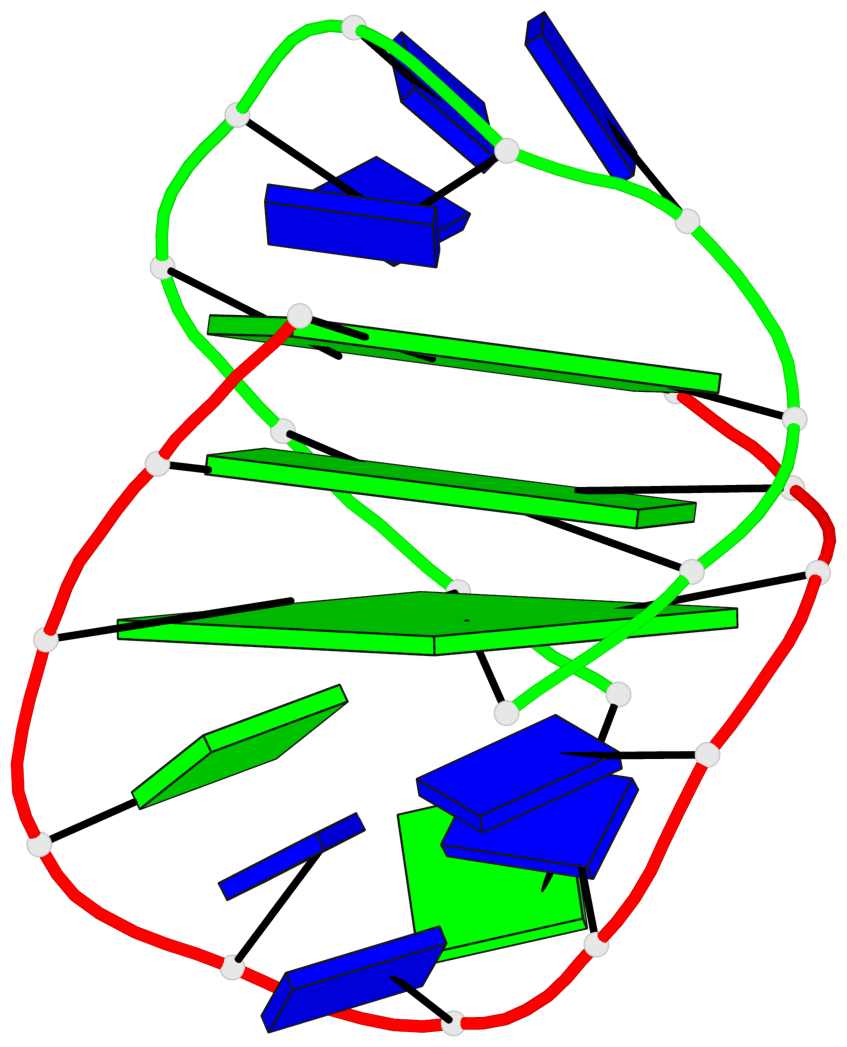
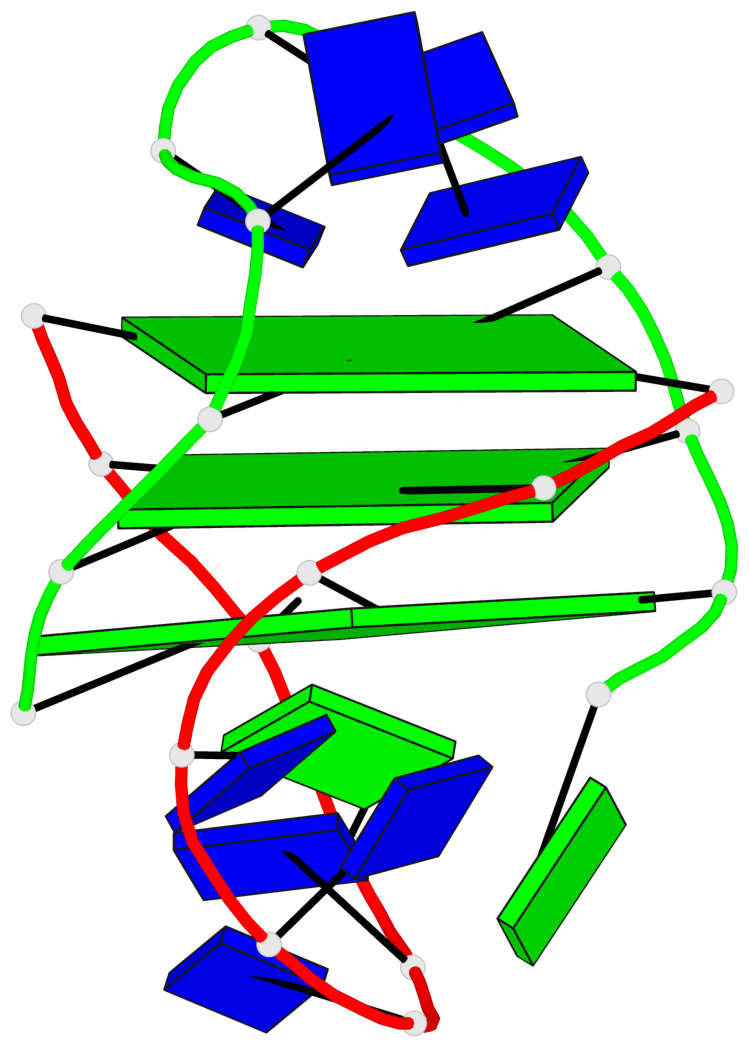
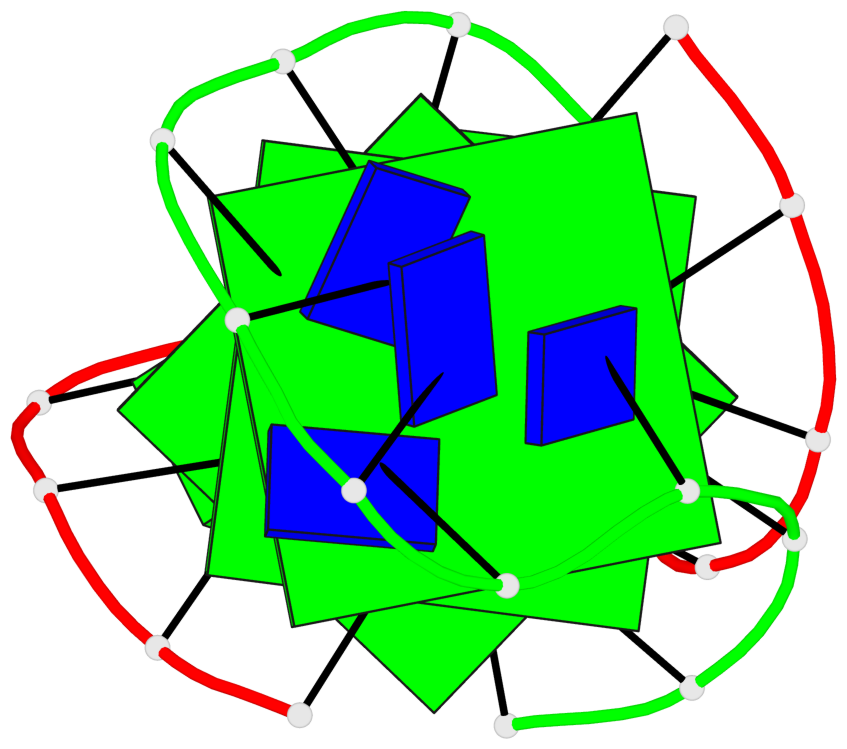
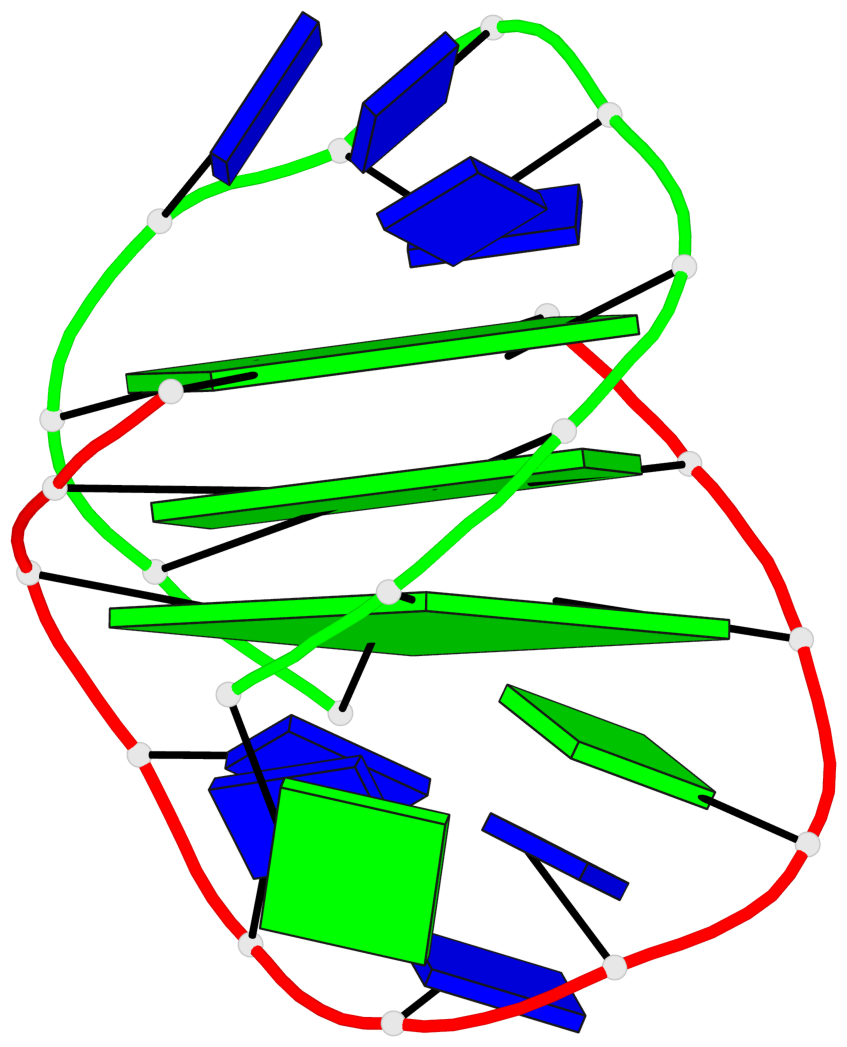
Detailed DSSR results for the G-quadruplex: PDB entry 1lvs
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 1lvs
- Class
- DNA
- Method
- NMR
- Summary
- The solution structure of d(g4t4g3)2
- Reference
- Crnugelj M, Hud NV, Plavec J (2002): "The solution structure of d(G(4)T(4)G(3))(2): a bimolecular G-quadruplex with a novel fold." J.Mol.Biol., 320, 911-924. doi: 10.1016/S0022-2836(02)00569-7.
- Abstract
- The G-rich 11-mer oligonucleotide d(G(4)T(4)G(3)) forms a bimolecular G-quadruplex in the presence of sodium ions with a topology that is distinct from the folds of the closely related and well-characterized sequences d(G(4)T(4)G(4)) and d(G(3)T(4)G(3)). The solution structure of d(G(4)T(4)G(3))(2) has been determined using a combination of NMR spectroscopy and restrained molecular dynamics calculations. d(G(4)T(4)G(3))(2) forms an asymmetric dimeric fold-back structure consisting of three stacked G-quartets. The two T(4) loops that span diagonally across the outer faces of the G-quartets assume different conformations. The glycosidic torsion angle conformations of the guanine bases are 5'-syn-anti-syn-anti-(T(4) loop)-anti-syn-anti in one strand and 5'-syn-anti-syn-anti-(T(4) loop)-syn-anti-syn in the other strand. The guanine bases of the two outer G-quartets exhibit a clockwise donor-acceptor hydrogen-bonding directionality, while those of the middle G-quartet exhibit the anti-clockwise directionality. The topology of this G-quadruplex, like other bimolecular fold-back structures with diagonal loops, places each strand of the G-quartet region next to a neighboring parallel and an anti-parallel strand. The two guanine residues not involved in G-quartet formation, G4 and G12 (i.e. the fourth guanine base of one strand and the first guanine base of the other strand), adopt distinct conformations. G4 is stacked on top of an adjacent G-quartet, and this base-stacking continues along with the bases of the loop residues T5 and T6. G12 is orientated away from the core of G-quartets; stacked on the T7 base and apparently involved in hydrogen-bonding interactions with the phosphodiester group of this same residue. The cation-dependent folding of the d(G(4)T(4)G(3))(2) quadruplex structure is distinct from that observed for similar sequences. While both d(G(4)T(4)G(4)) and d(G(3)T(4)G(3)) form bimolecular, diagonally looped G-quadruplex structures in the presence of Na(+), K(+) and NH(4)(+), we have observed this folding to be favored for d(G(4)T(4)G(3)) in the presence of Na(+), but not in the presence of K(+) or NH(4)(+). The structure of d(G(4)T(4)G(3))(2) exhibits a "slipped-loop" element that is similar to what has been proposed for structural intermediates in the folding pathway of some G-quadruplexes, and therefore provides support for the feasibility of these proposed transient structures in G-quadruplex formation.
- G4 notes
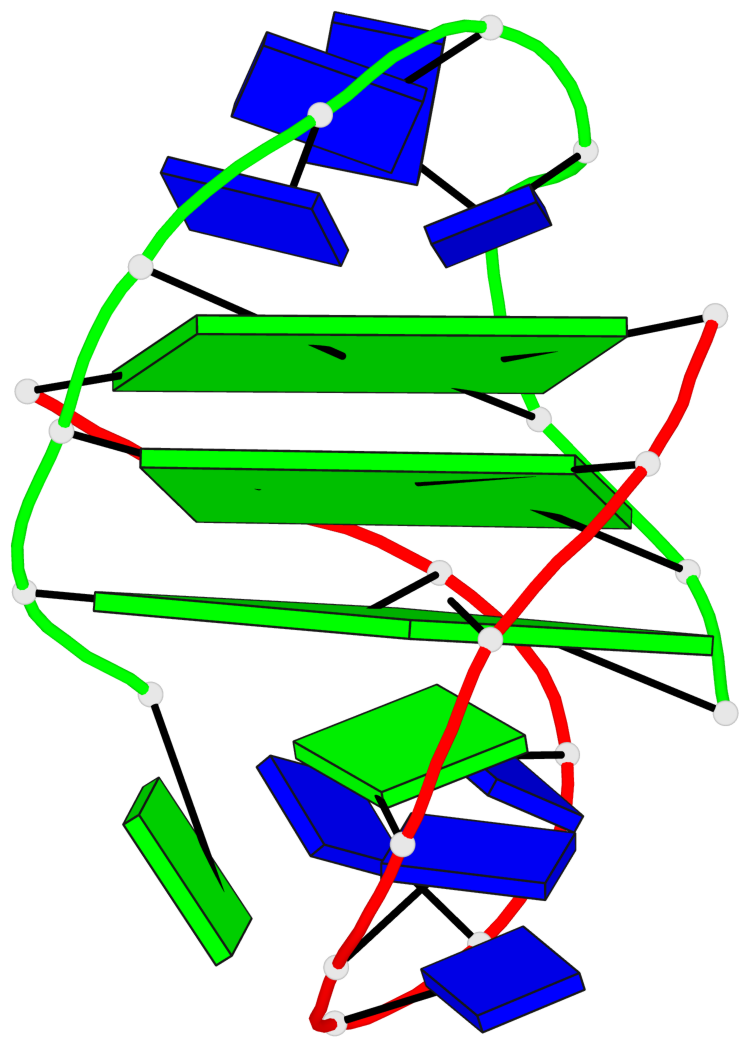
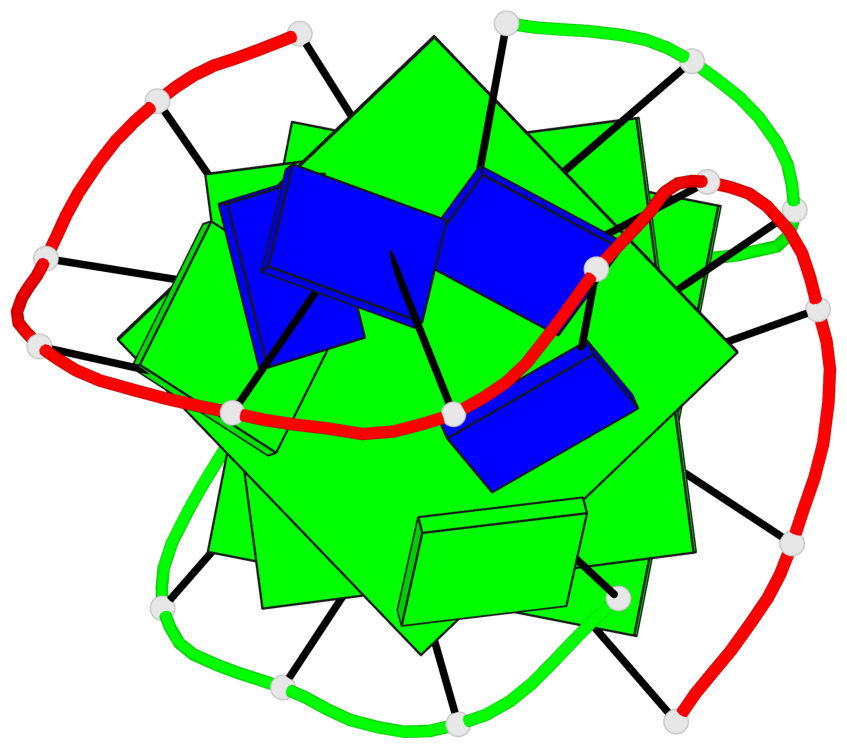
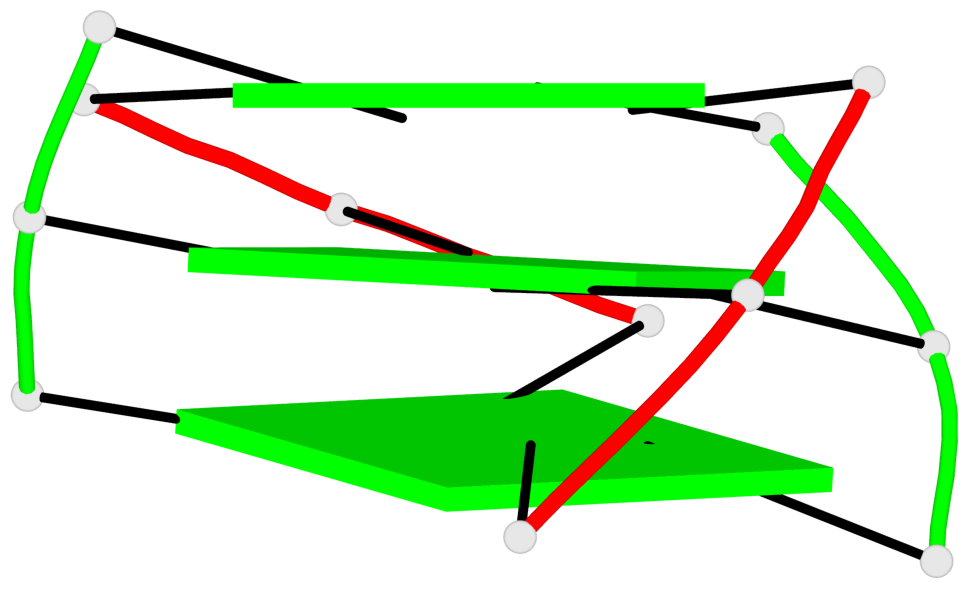
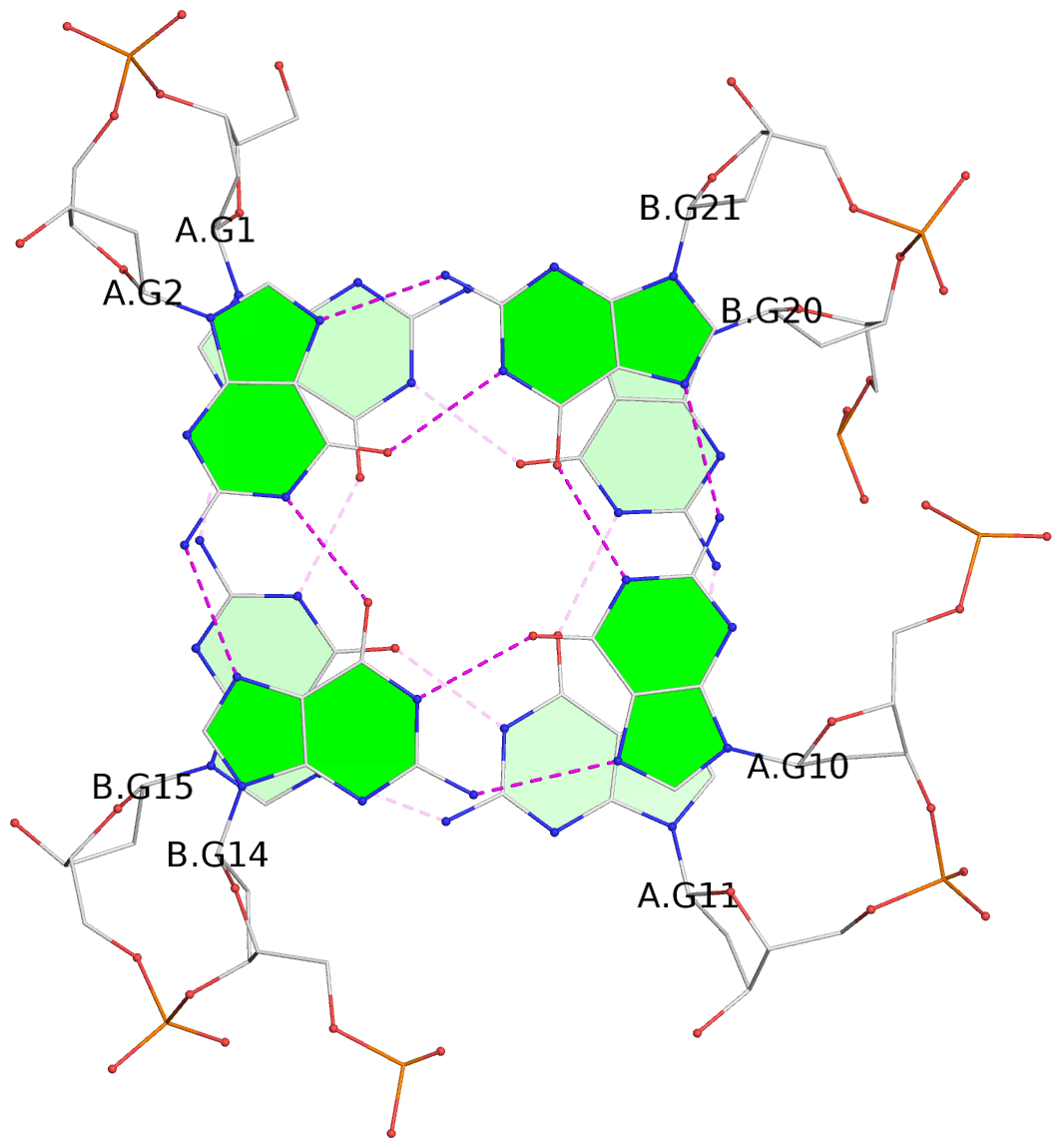
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, (2+2), UDDU
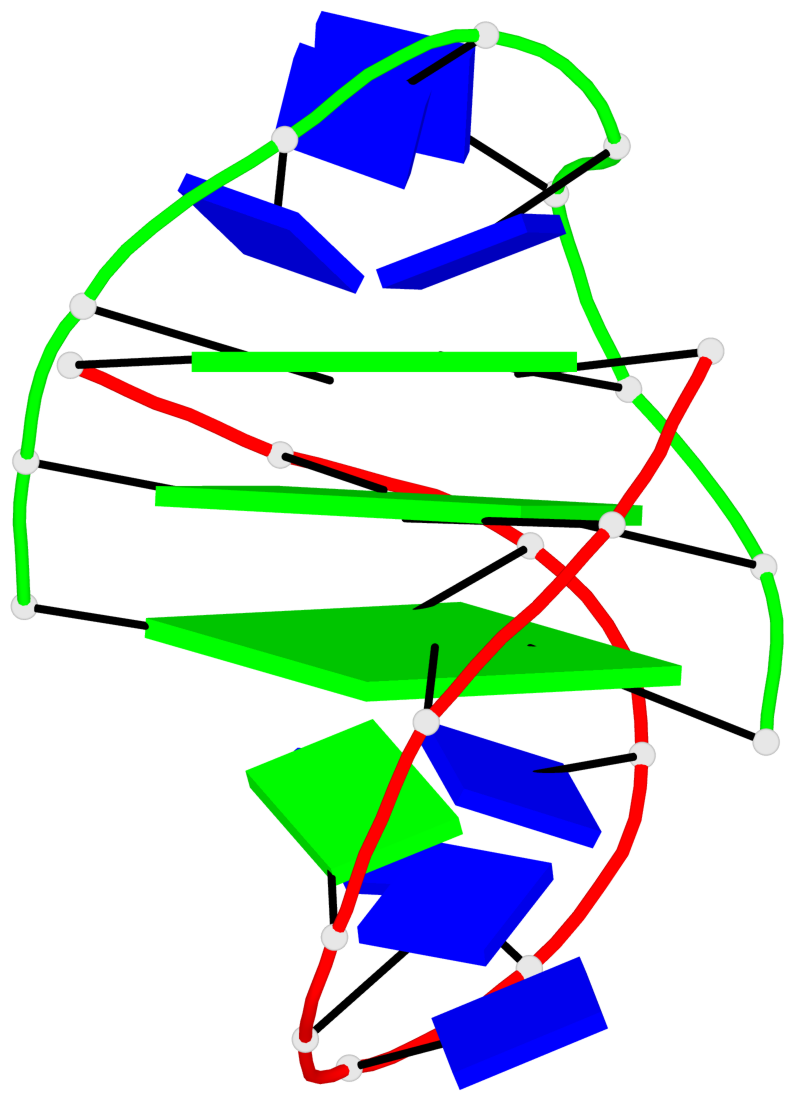
Base-block schematics in six views
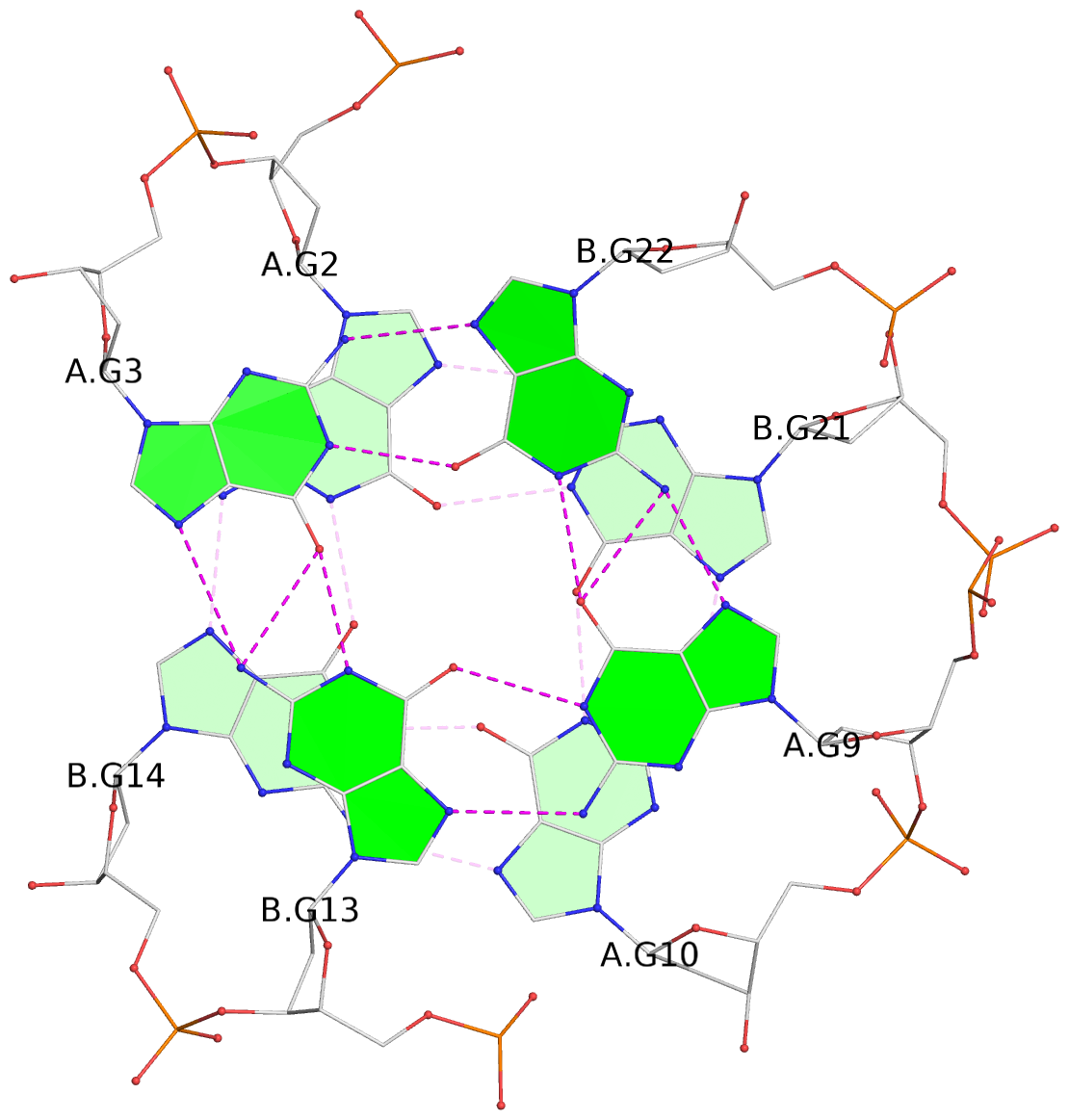
List of 3 G-tetrads
1 glyco-bond=s--s sugar=---- groove=w-n- planarity=0.332 type=other nts=4 GGGG A.DG1,B.DG15,A.DG11,B.DG20 2 glyco-bond=-ss- sugar=---- groove=w-n- planarity=0.227 type=other nts=4 GGGG A.DG2,B.DG14,A.DG10,B.DG21 3 glyco-bond=s--s sugar=---- groove=w-n- planarity=0.302 type=other nts=4 GGGG A.DG3,B.DG13,A.DG9,B.DG22
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, inter-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.