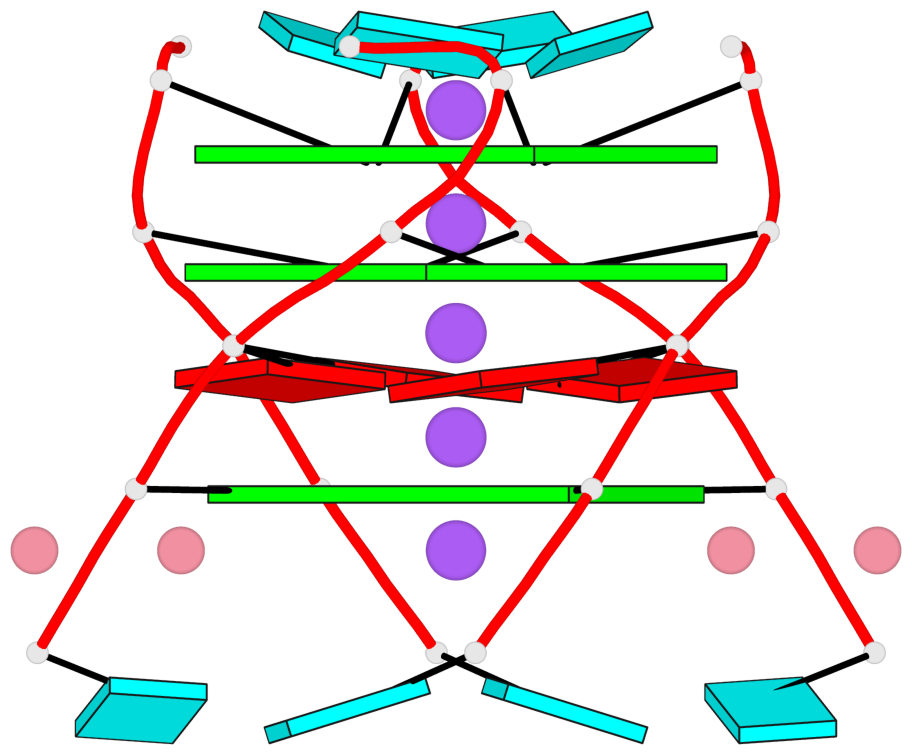
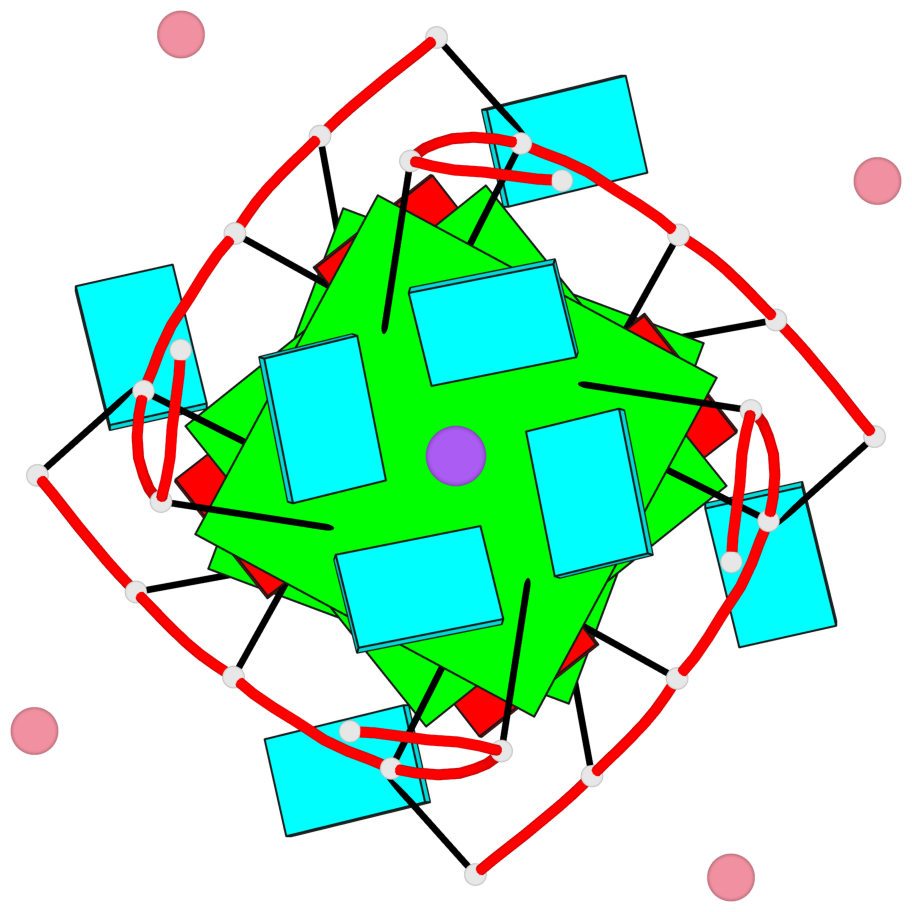
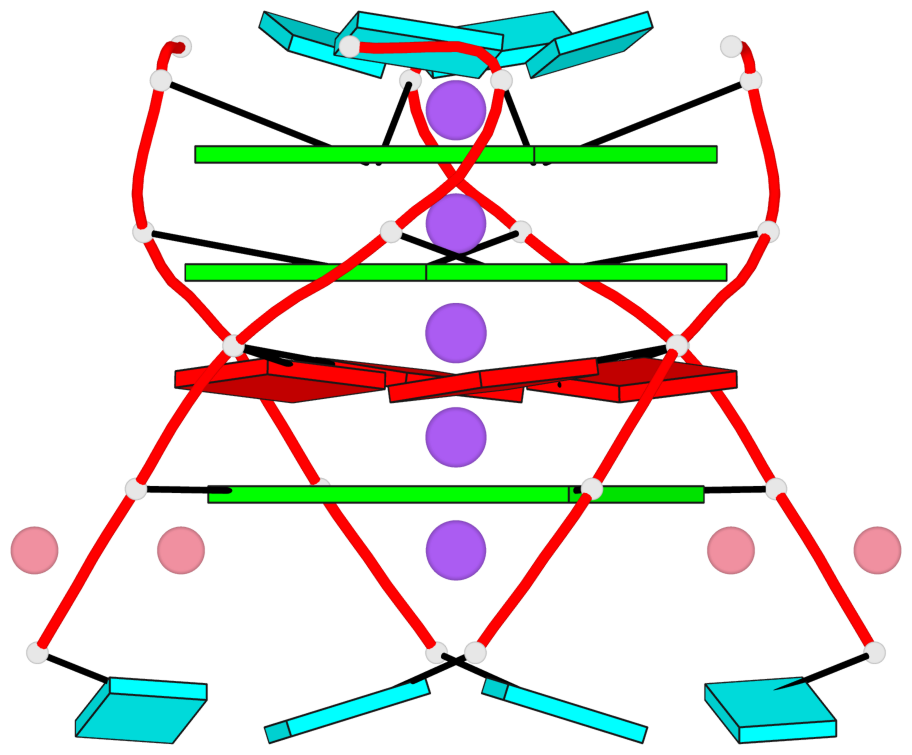
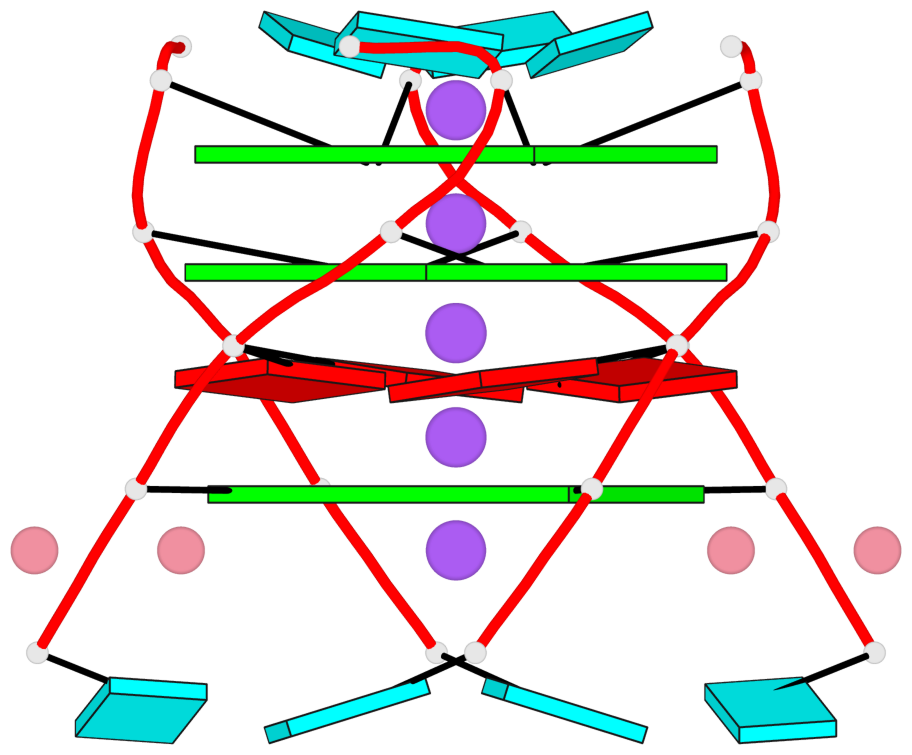
Detailed DSSR results for the G-quadruplex: PDB entry 1mdg
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 1mdg
- Class
- RNA
- Method
- X-ray (1.5 Å)
- Summary
- An alternating antiparallel octaplex in an RNA crystal structure
- Reference
- Pan BC, Xiong Y, Shi K, Sundaralingam M (2003): "An Eight-Stranded Helical Fragment in RNA Crystal Structure: Implications for Tetraplex Interaction." Structure, 11, 825-831. doi: 10.1016/S0969-2126(03)00108-4.
- Abstract
- Multistranded helical structures in nucleic acids play various functions in biological processes. Here we report the crystal structure of a hexamer, rU(BrdG)r(AGGU),at 1.5 A resolution containing a structural complex of an alternating antiparallel eight-stranded helical fragment that is sandwiched in two tetraplexes. The octaplex is formed by groove binding interaction and base tetrad intercalation between two tetraplexes. Two different forms of octaplexes have been proposed, which display different properties in interaction with proteins and nucleic acids. Adenines form a base tetrad in the novel N6-H em leader N3 conformation and further interact with uridines to form an adenine-uridine octad in the reverse Hoogsteen pairing scheme. The conformational flexibility of adenine tetrad indicates that it can optimize its conformation in different interactions.
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, parallel(4+0), UUUU
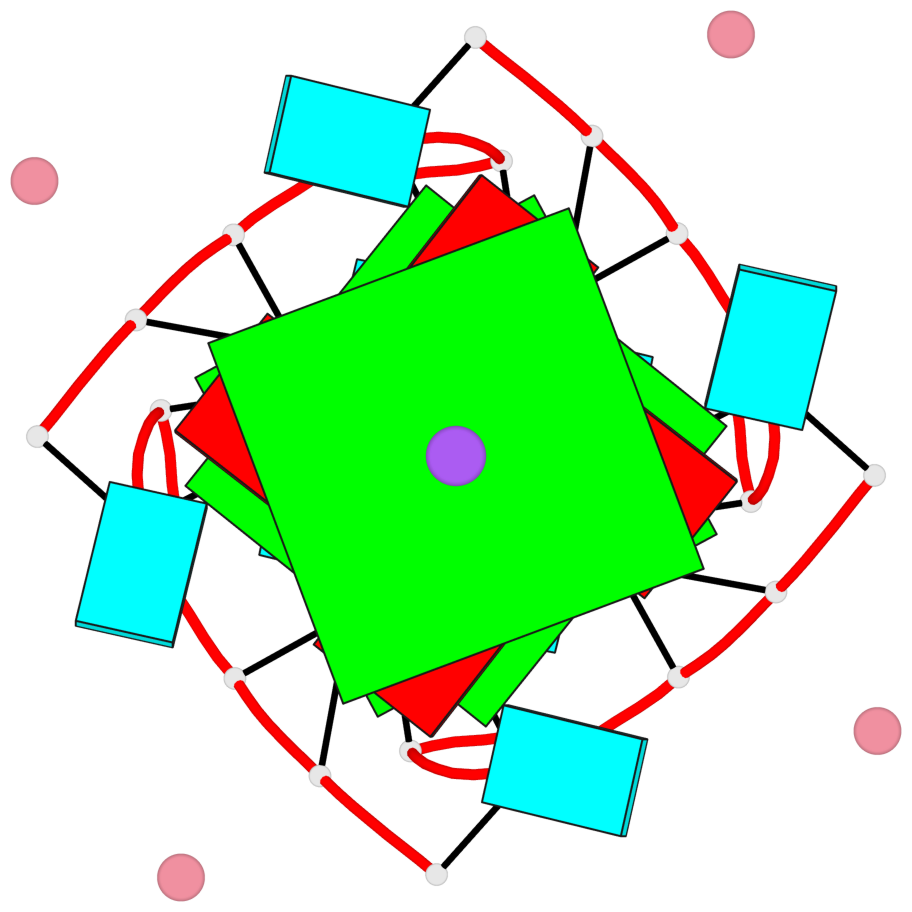
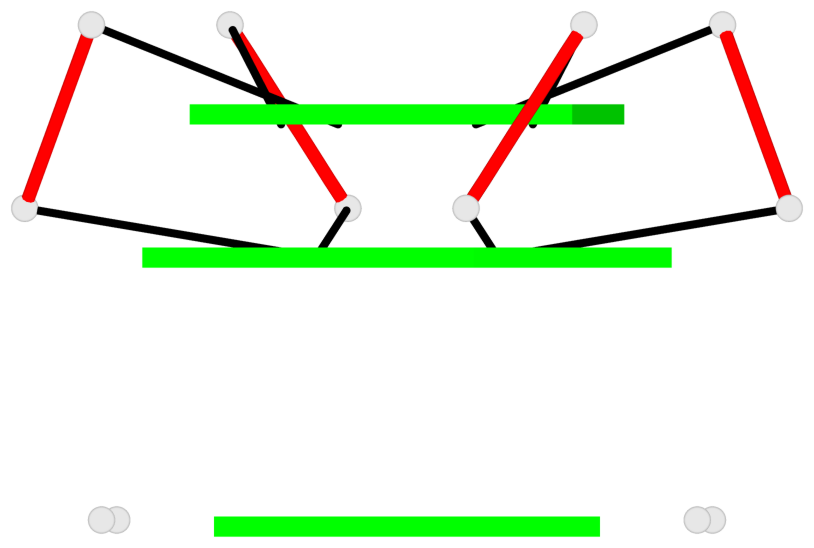
Base-block schematics in six views
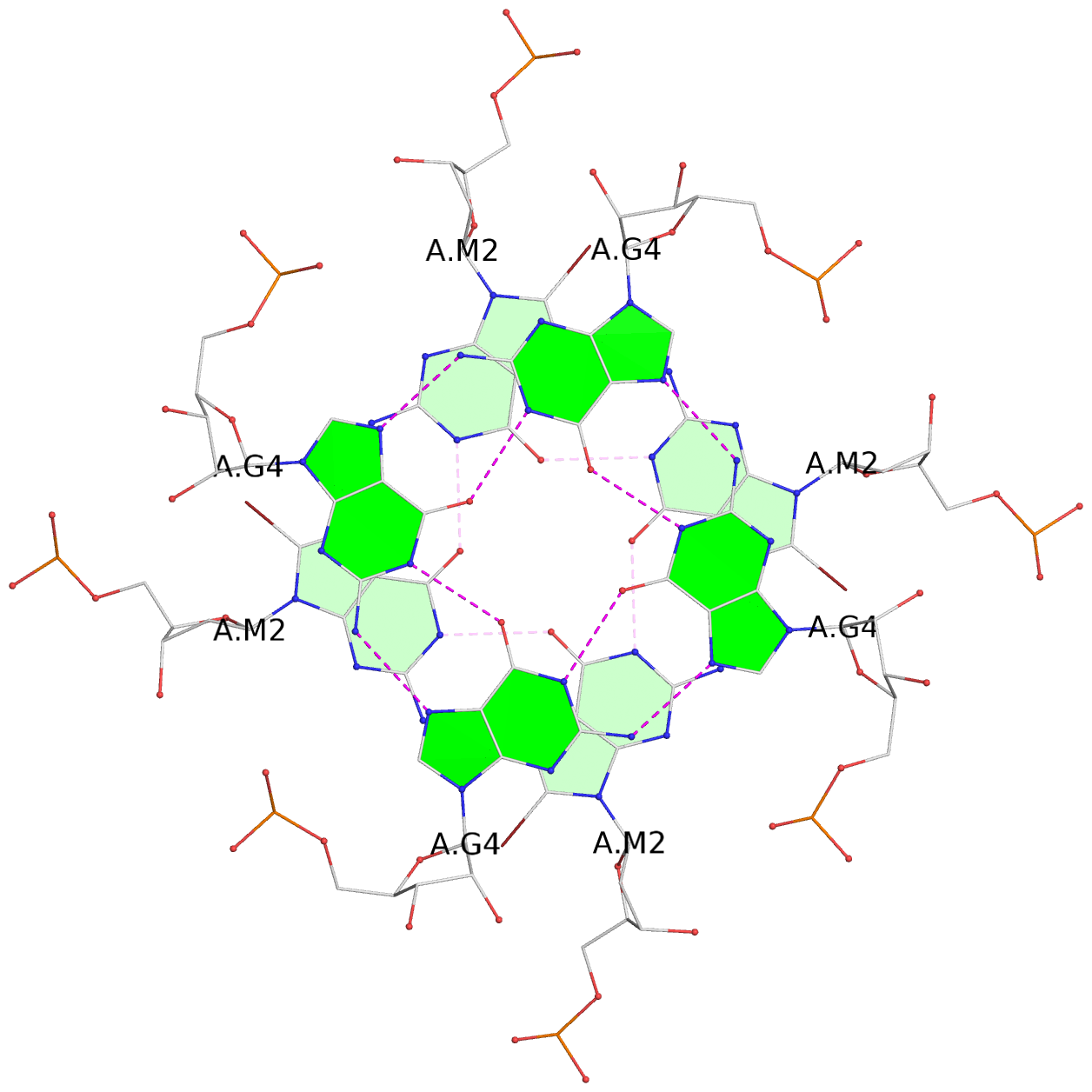
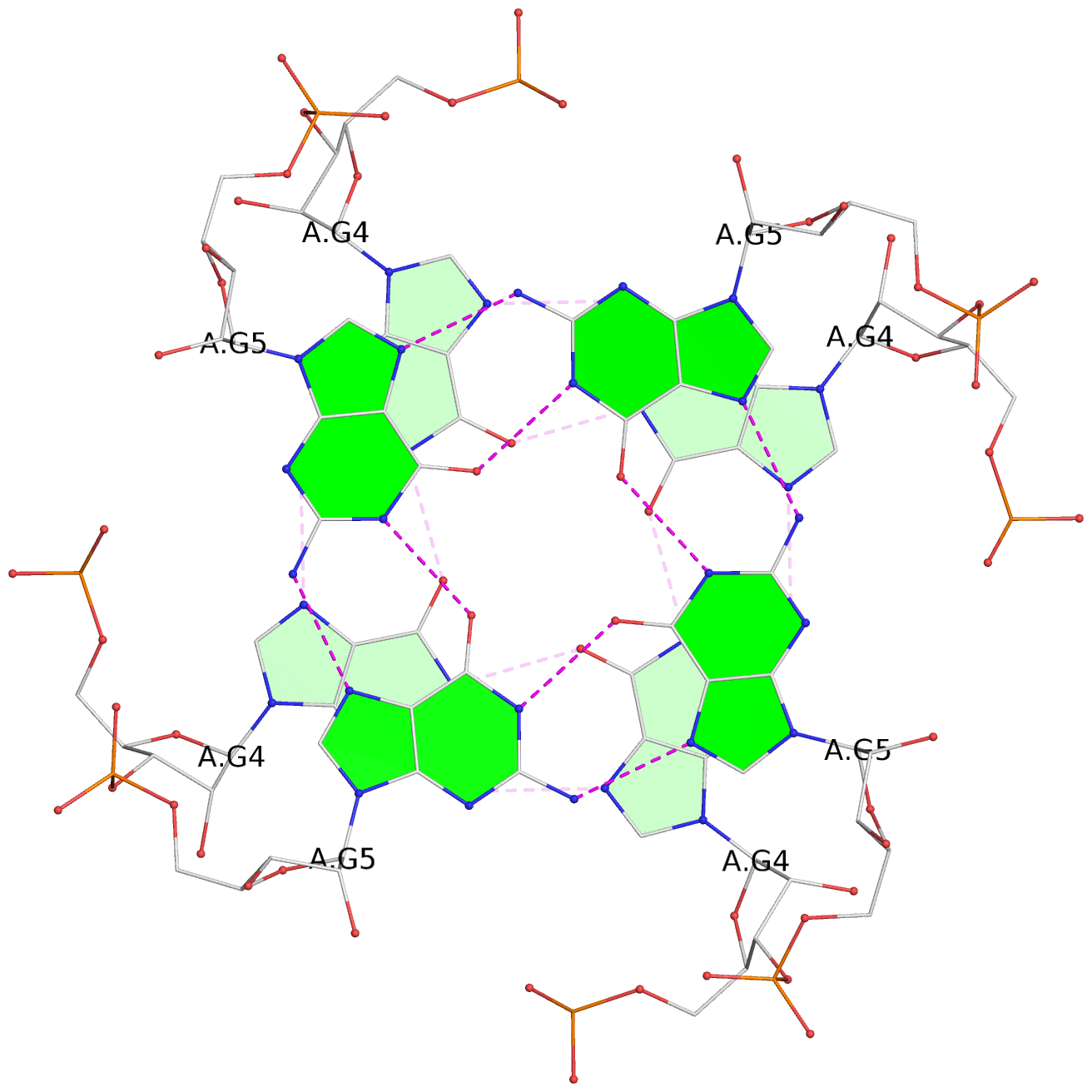
List of 3 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.110 type=planar nts=4 gggg 1:A.BGM2,4:A.BGM2,2:A.BGM2,3:A.BGM2 2 glyco-bond=---- sugar=3333 groove=---- planarity=0.171 type=other nts=4 GGGG 1:A.G4,4:A.G4,2:A.G4,3:A.G4 3 glyco-bond=---- sugar=3333 groove=---- planarity=0.230 type=bowl nts=4 GGGG 1:A.G5,4:A.G5,2:A.G5,3:A.G5
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, inter-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.