Detailed DSSR results for the G-quadruplex: PDB entry 1my9
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 1my9
- Class
- RNA
- Method
- NMR
- Summary
- Solution structure of a k+ cation stabilized dimeric RNA quadruplex containing two g:g(:a):g:g(:a) hexads, g:g:g:g tetrads and uuuu loops
- Reference
- Liu H, Matsugami A, Katahira M, Uesugi S (2002): "A Dimeric RNA Quadruplex Architecture Comprised of Two G:G(:A):G:G(:A) Hexads, G:G:G:G Tetrads and UUUU Loops." J.Mol.Biol., 322, 955-970. doi: 10.1016/S0022-2836(02)00876-8.
- Abstract
- Using CD and NMR, we determined the structure of an RNA oligomer, r(GGAGGUUUUGGAGG) (R14), comprising two GGAGG segments joined by a UUUU segment. A modified quadruplex structure was observed for r(GGAGGUUUUGGAGG) in solution even in the absence of K(+). An unusually stable dimeric RNA quadruplex architecture formed from two strands of r(GGAGGUUUUGGAGG) at low K(+) concentration is reported here. In each strand of r(GGAGGUUUUGGAGG), two sets of successive turns in the GGAGG segments and turns at both ends of the UUUU loops drive four G-G steps to align in a parallel manner, a core with two stacked G-tetrads being formed. Two adenine bases bind to two edges of one G:G:G:G tetrad through the sheared G:A mismatch augmenting the tetrad into a G:G(:A):G:G(:A) hexad. Thus, one molecule of r(GGAGGUUUUGGAGG) folds into a modified quadruplex comprising a G:G:G:G tetrad, a UUUU double-chain reversal loop and a G:G(:A):G:G(:A) hexad. Two such molecules further associate by stacking through the dimeric hexad-hexad interface with a rotational symmetry. The ribose rings of most nucleotides take S (close to C2'-endo) puckering, which is unusual for an RNA. K(+) can increase the stability of this quadruplex structure; the number of bound K(+) was estimated from the results of the titration experiment. Besides G:G and G:A mismatches, a network of hydrogen bonds including O4'-NH(2) and C-H..O hydrogen bonds, and the extensive base stacking contribute to the high thermodynamic stability of R14. Our results could provide the stereochemical and thermodynamic basis for elucidating the biological role of the GGAGG-containing RNA segments abundantly existing in various RNAs. Relevance to quadruplex-mediated mRNA-FMRP binding and HIV-1 genome RNA dimerization is discussed.
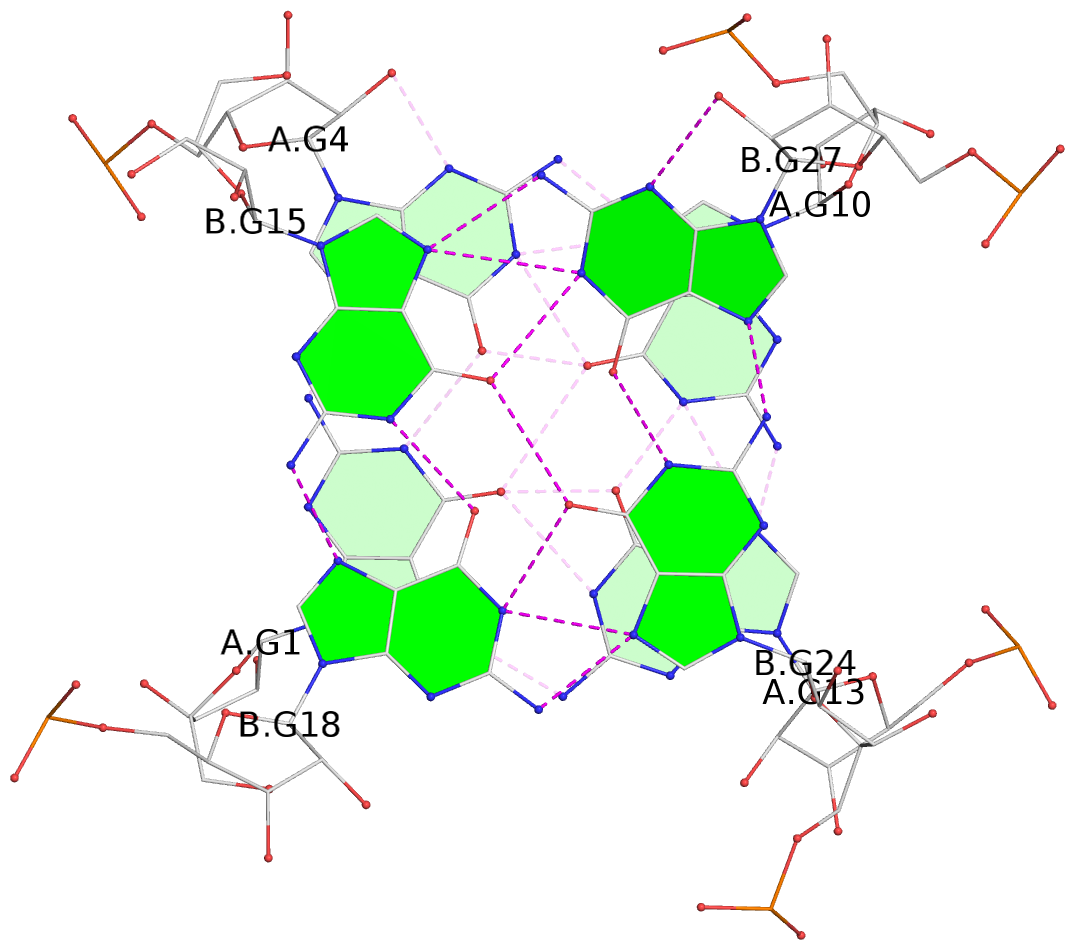
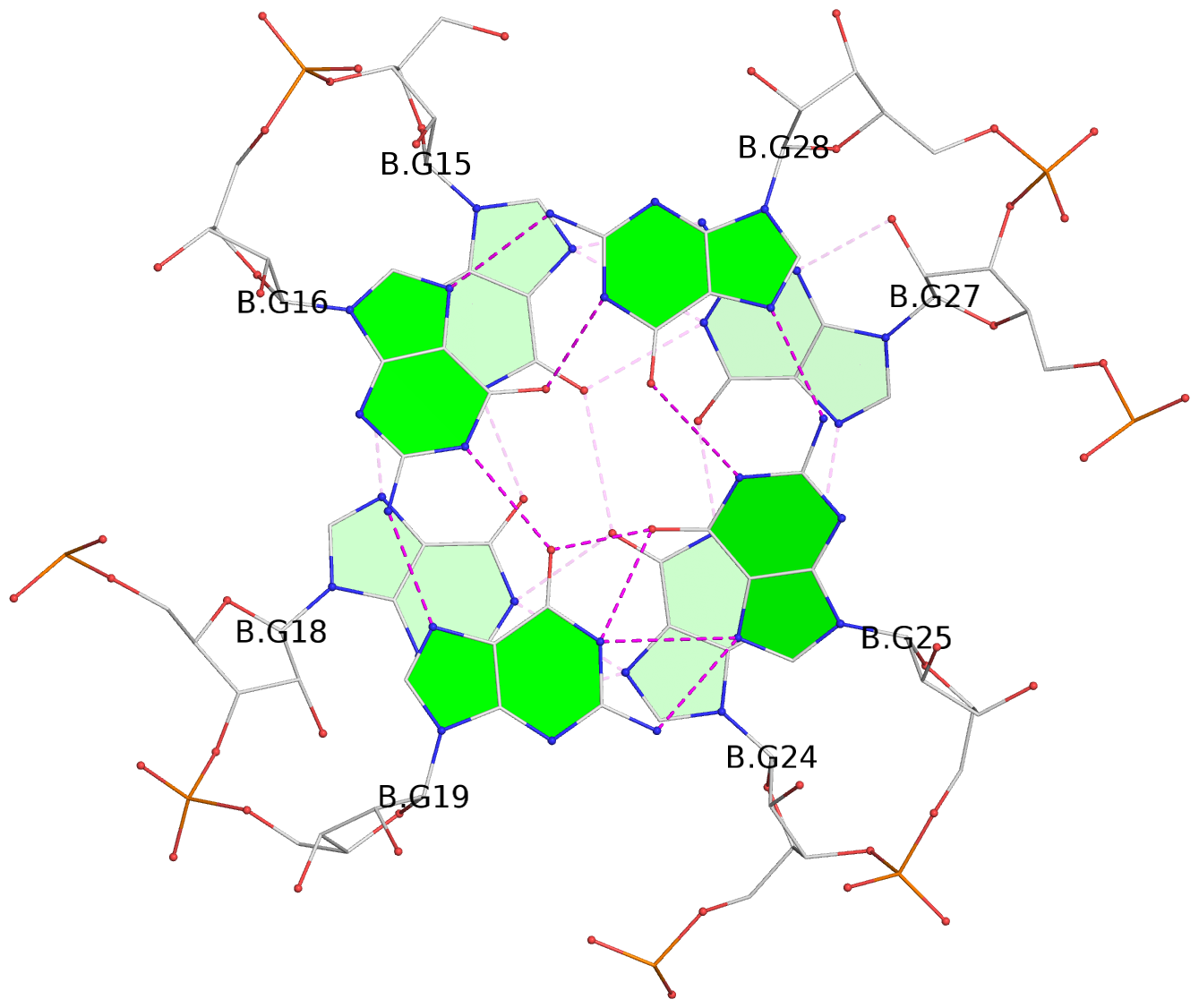
- G4 notes
- 4 G-tetrads, 1 G4 helix, 2 G4 stems, 1 G4 coaxial stack, 2(-P-P-P), parallel(4+0), UUUU, coaxial interfaces: 5'/5'
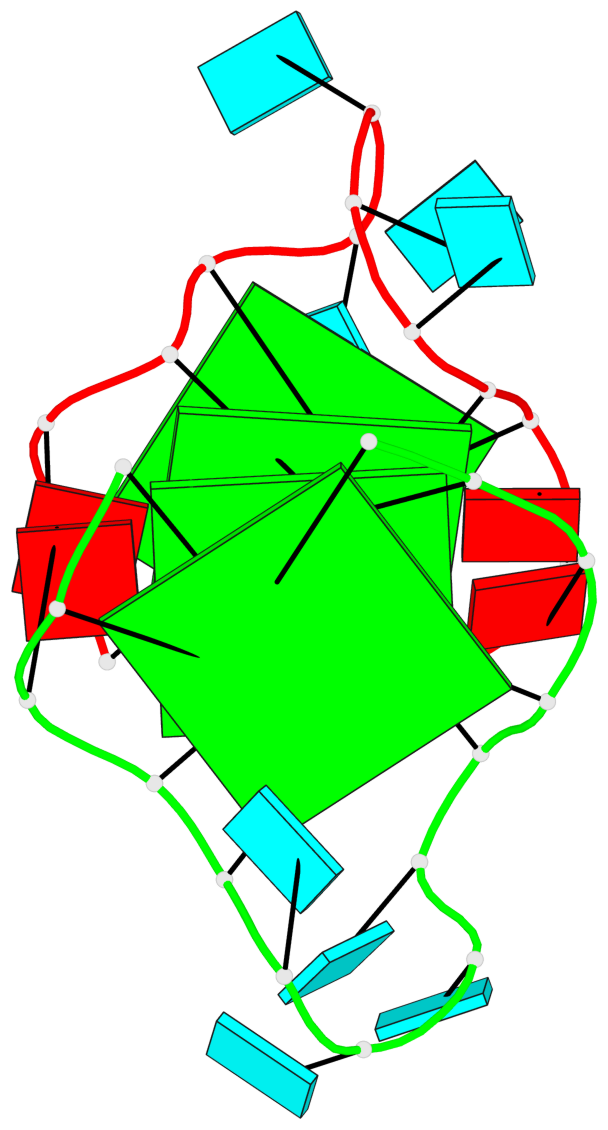
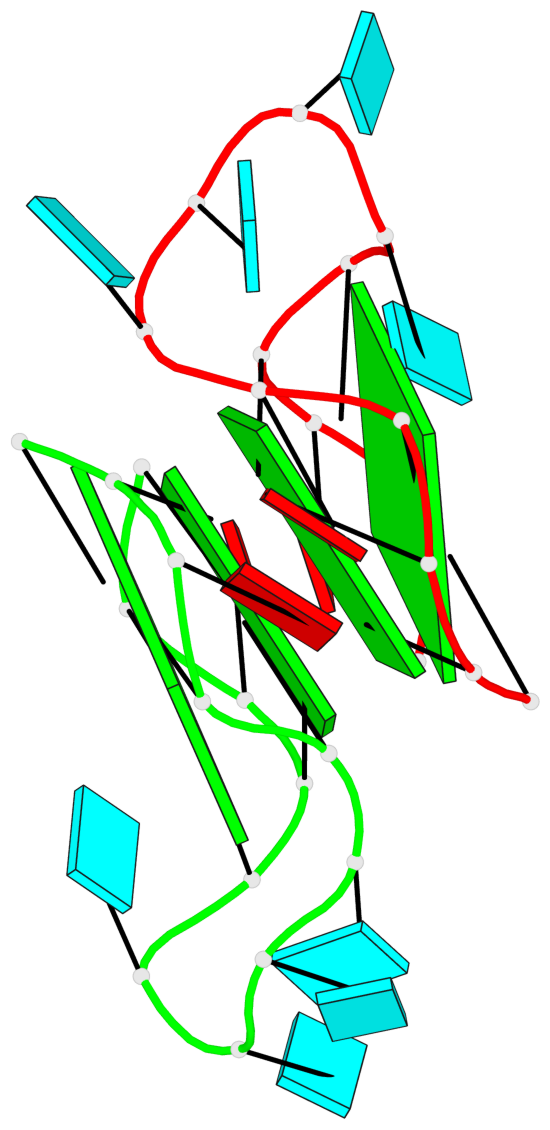
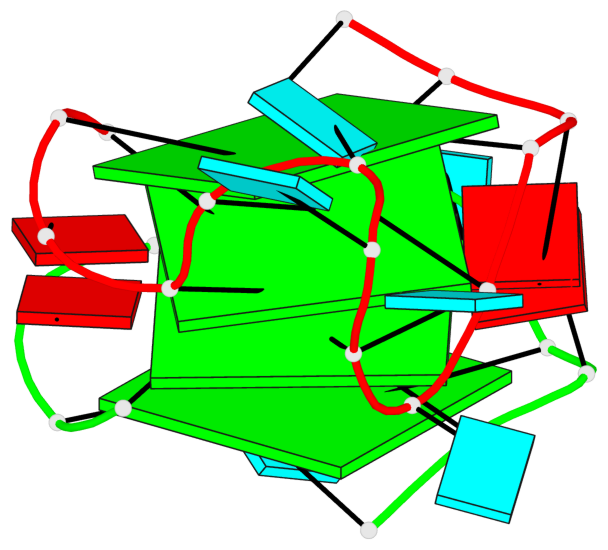
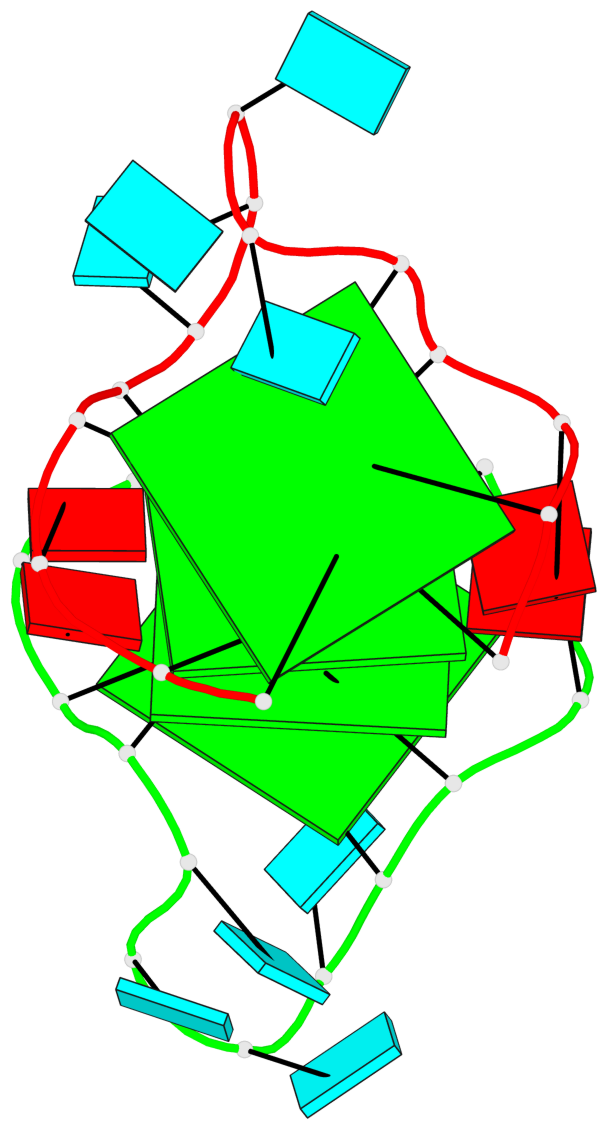
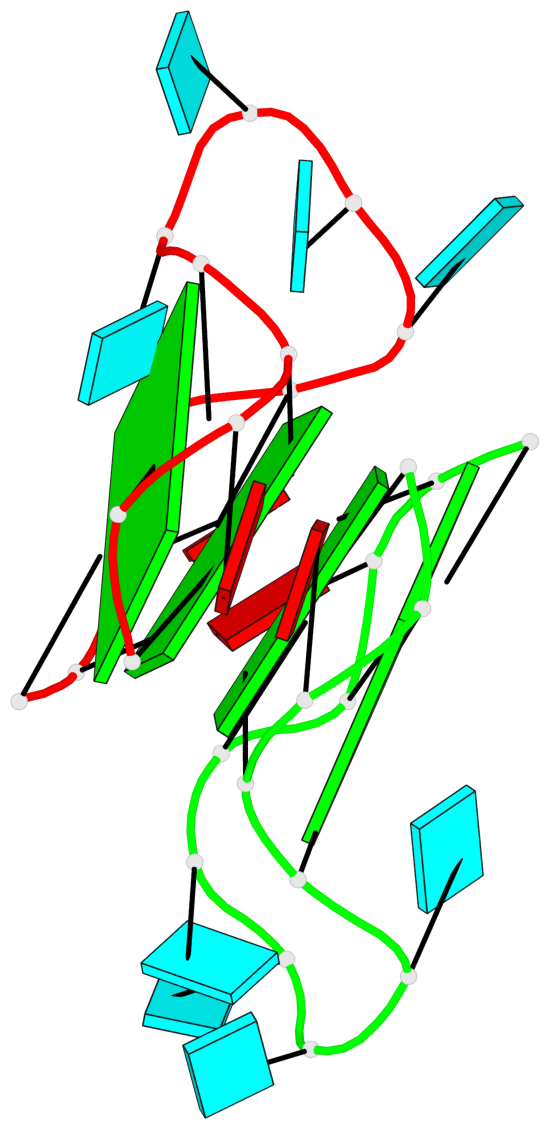
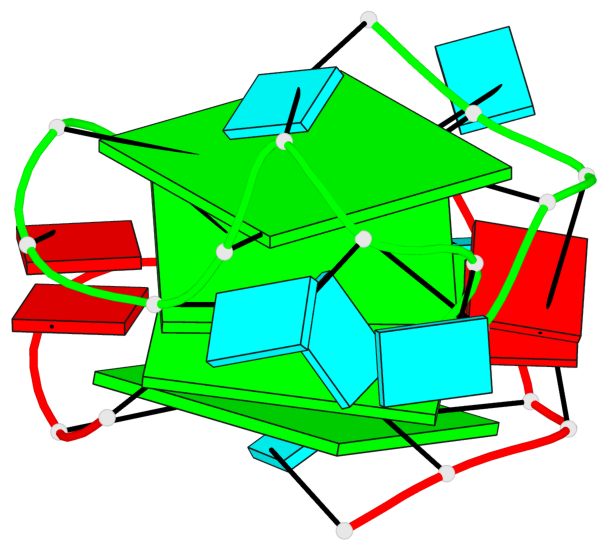
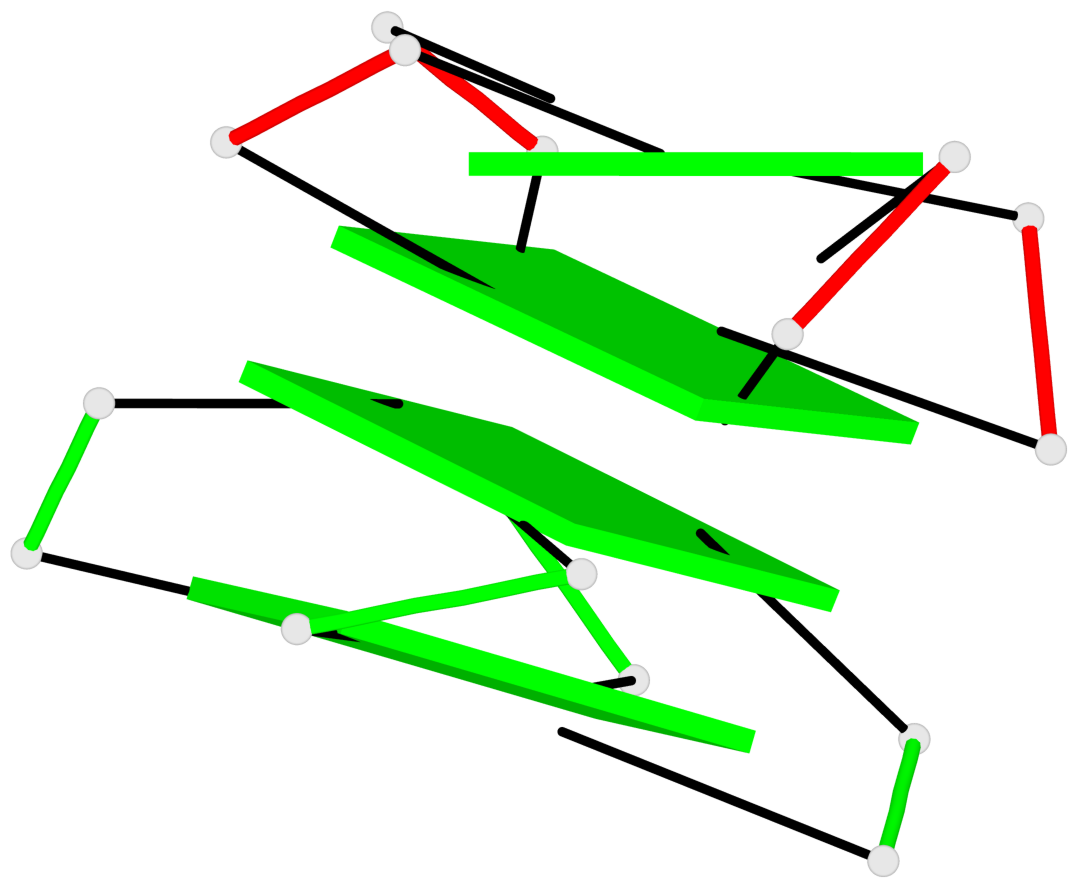
Base-block schematics in six views
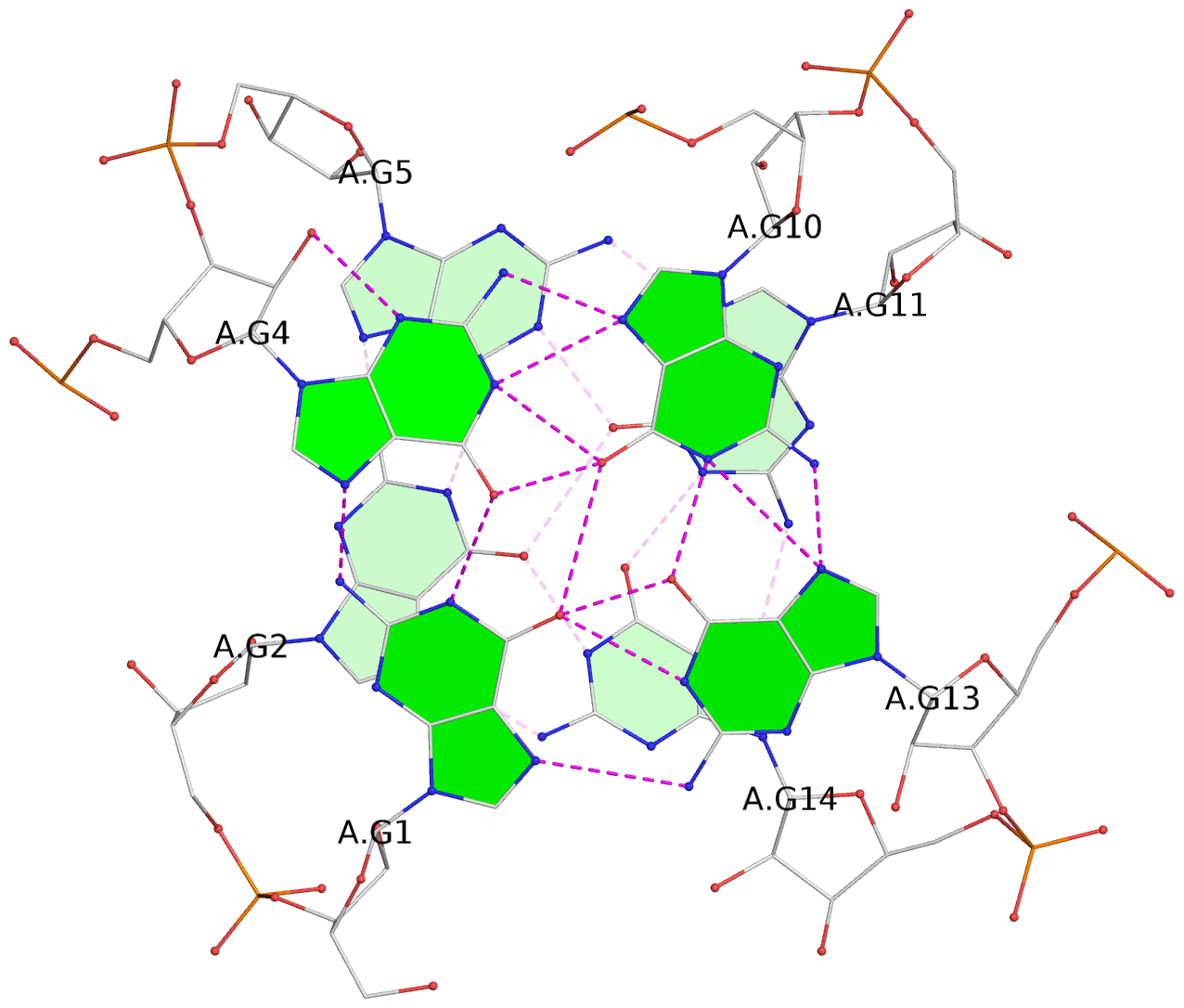
List of 4 G-tetrads
1 glyco-bond=---- sugar=---. groove=---- planarity=0.448 type=other nts=4 GGGG A.G1,A.G4,A.G10,A.G13 2 glyco-bond=---- sugar=-3-- groove=---- planarity=0.841 type=other nts=4 GGGG A.G2,A.G5,A.G11,A.G14 3 glyco-bond=---- sugar=-.-- groove=---- planarity=0.435 type=other nts=4 GGGG B.G15,B.G18,B.G24,B.G27 4 glyco-bond=---- sugar=-.-- groove=---- planarity=0.717 type=saddle nts=4 GGGG B.G16,B.G19,B.G25,B.G28
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 4 G-tetrad layers, inter-molecular, with 2 stems
List of 2 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 2 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 2(-P-P-P), parallel(4+0)
Stem#2, 2 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 2(-P-P-P), parallel(4+0)
List of 1 G4 coaxial stack
1 G4 helix#1 contains 2 G4 stems: [#1,#2] [5'/5']