Detailed DSSR results for the G-quadruplex: PDB entry 1n7b
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 1n7b
- Class
- DNA, RNA
- Method
- X-ray (1.4 Å)
- Summary
- Rip-radiation-damage induced phasing
- Reference
- Ravelli RBG, Leiros H-KS, Pan B, Caffrey M, McSweeney S (2003): "Specific Radiation-Damage Can Be Used To Solve Macromolecular Crystal Structures." Structure, 11, 217-224. doi: 10.1016/S0969-2126(03)00006-6.
- Abstract
- The use of third generation synchrotron sources has led to renewed concern about the effect of ionizing radiation on crystalline biological samples. In general, the problem is seen as one to be avoided. However, in this paper, it is shown that, far from being a hindrance to successful structure determination, radiation damage provides an opportunity for phasing macromolecular structures. This is successfully demonstrated for both a protein and an oligonucleotide, by way of which complete models were built automatically. The possibility that, through the exploitation of radiation damage, the phase problem could become less of a barrier to macromolecular crystal structure determination is discussed.
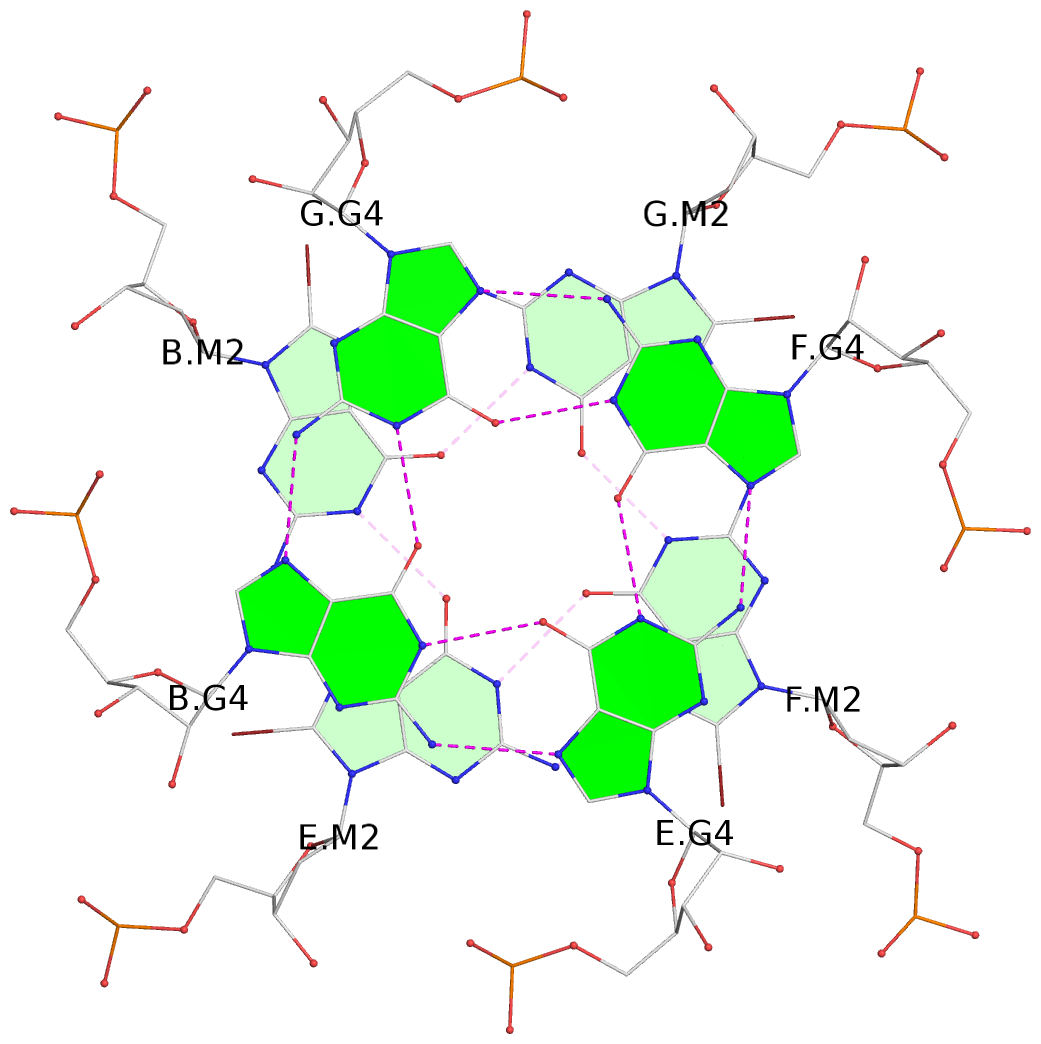
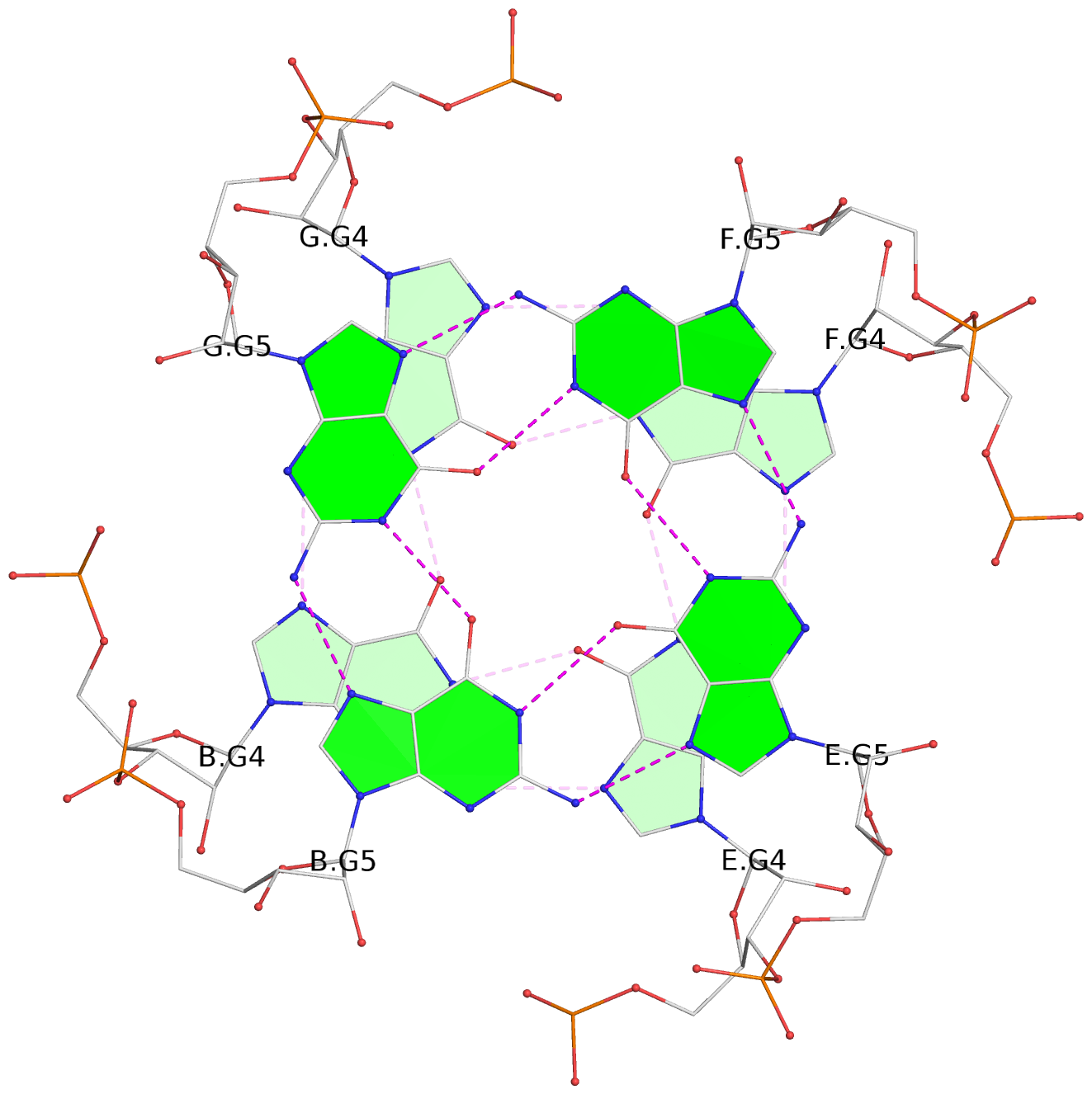
- G4 notes
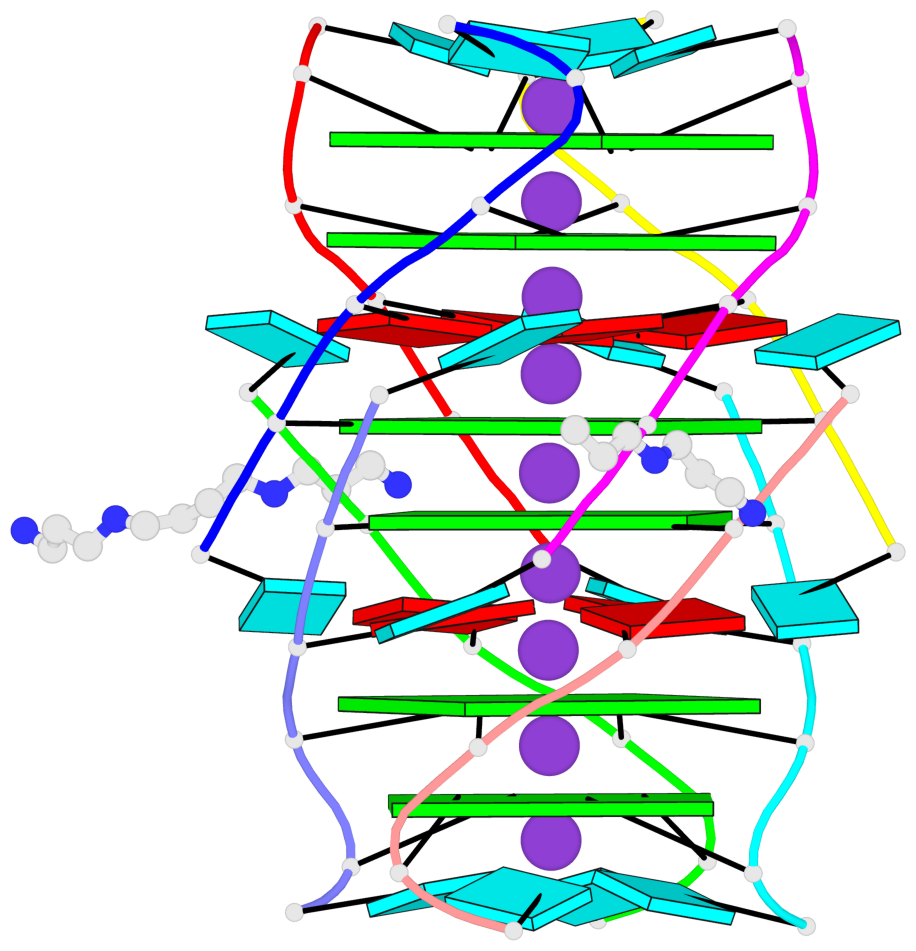
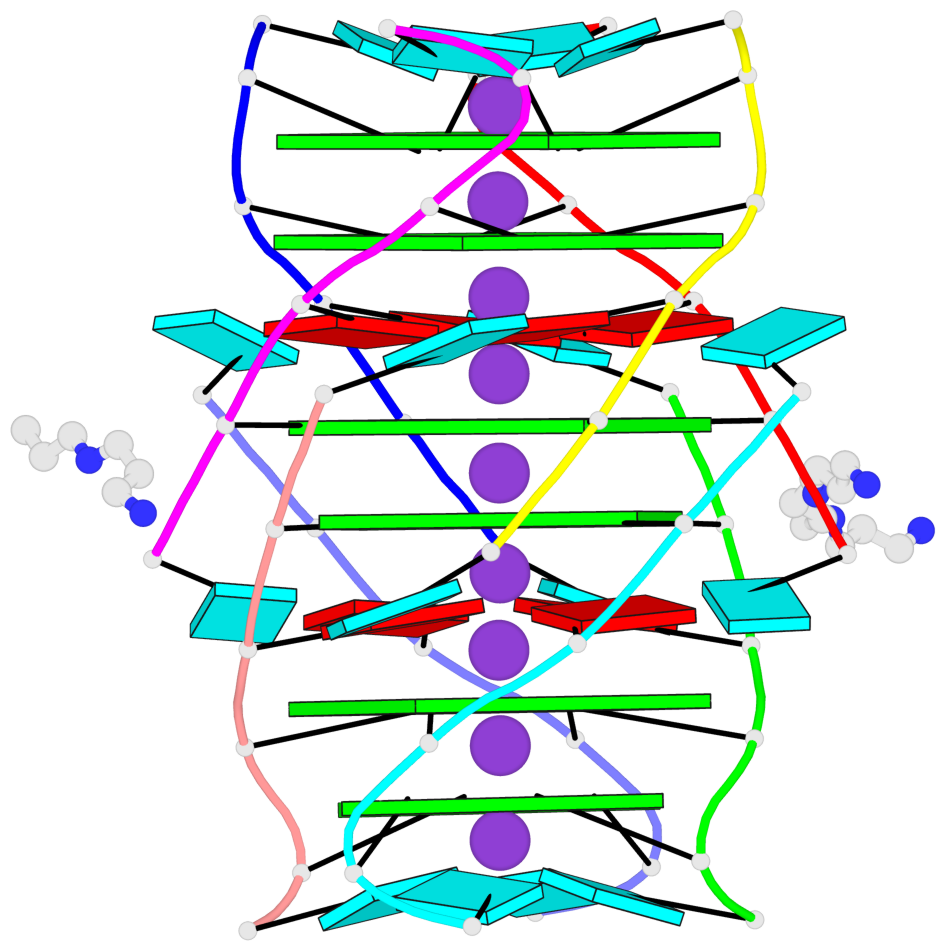
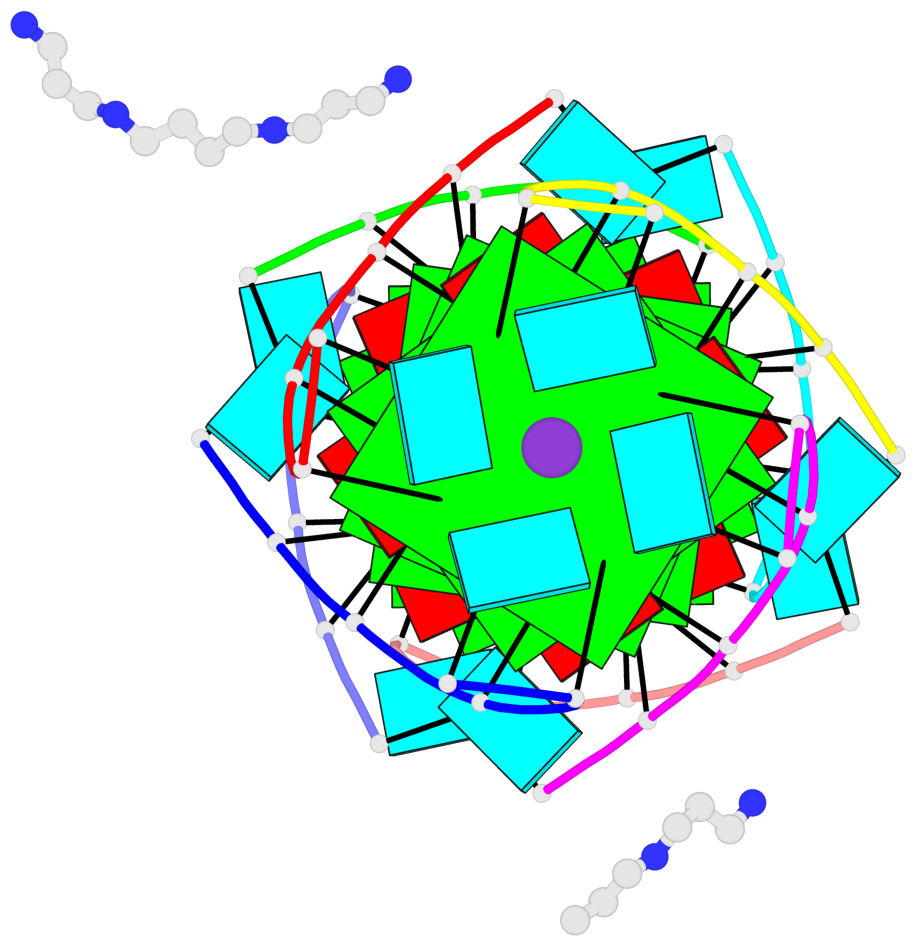
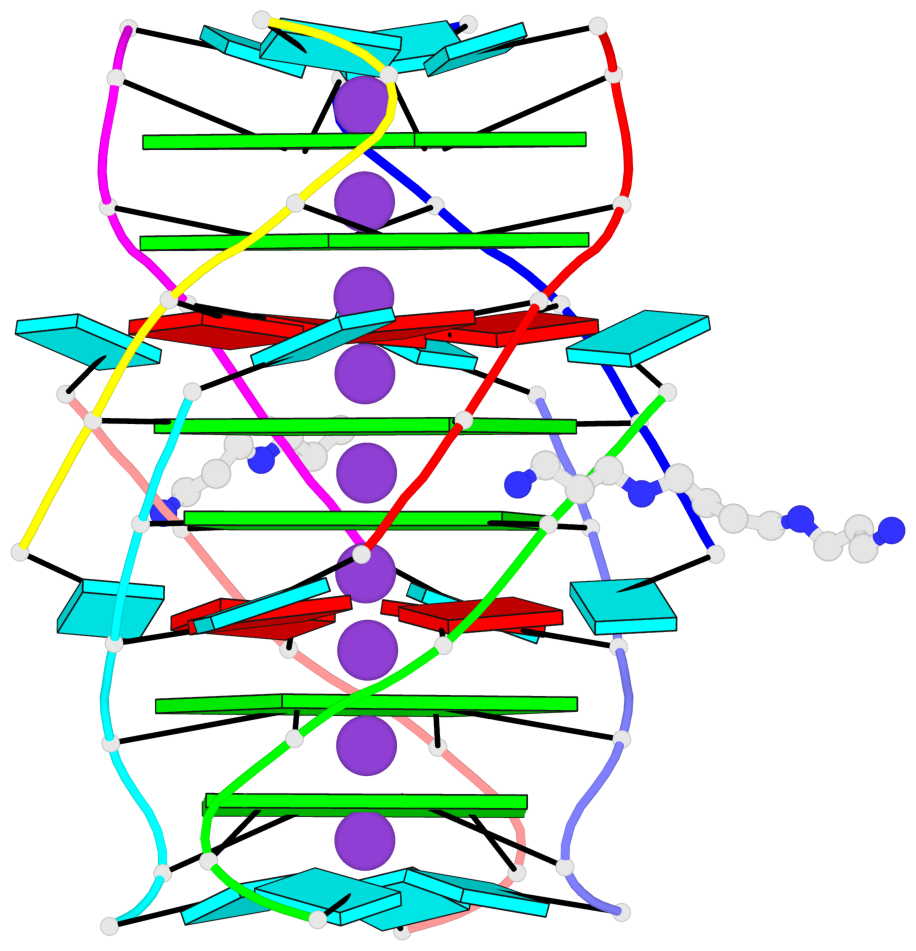
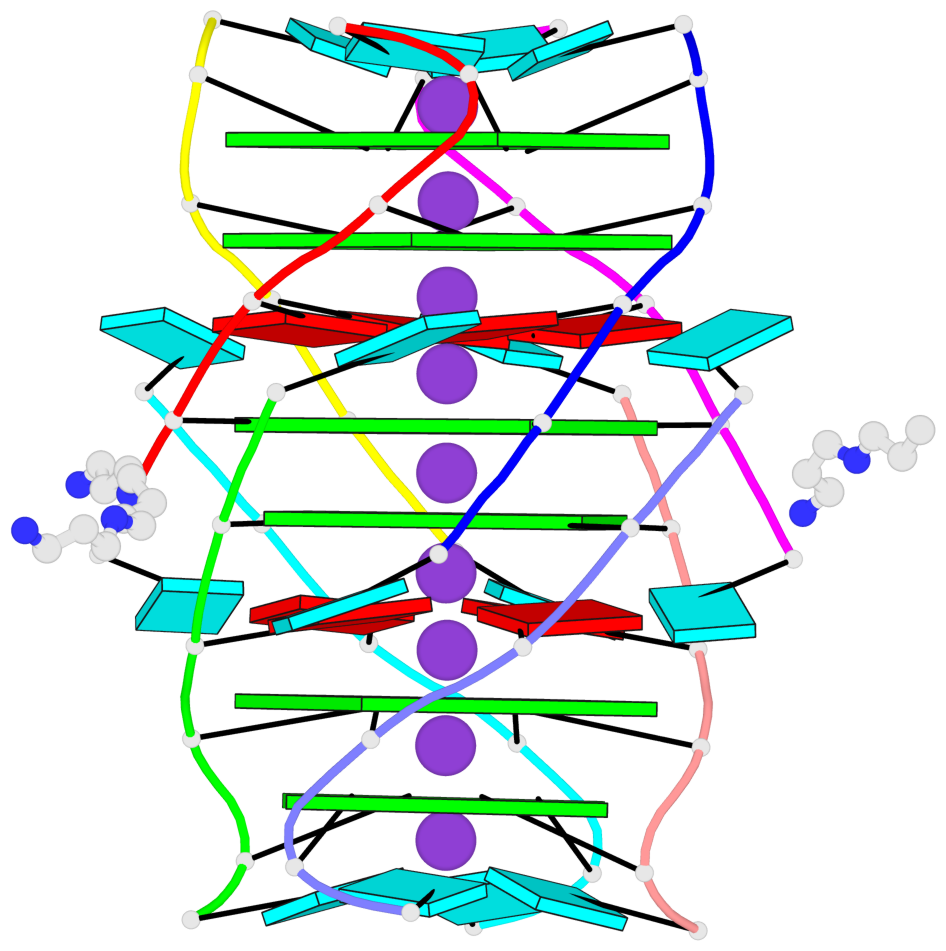
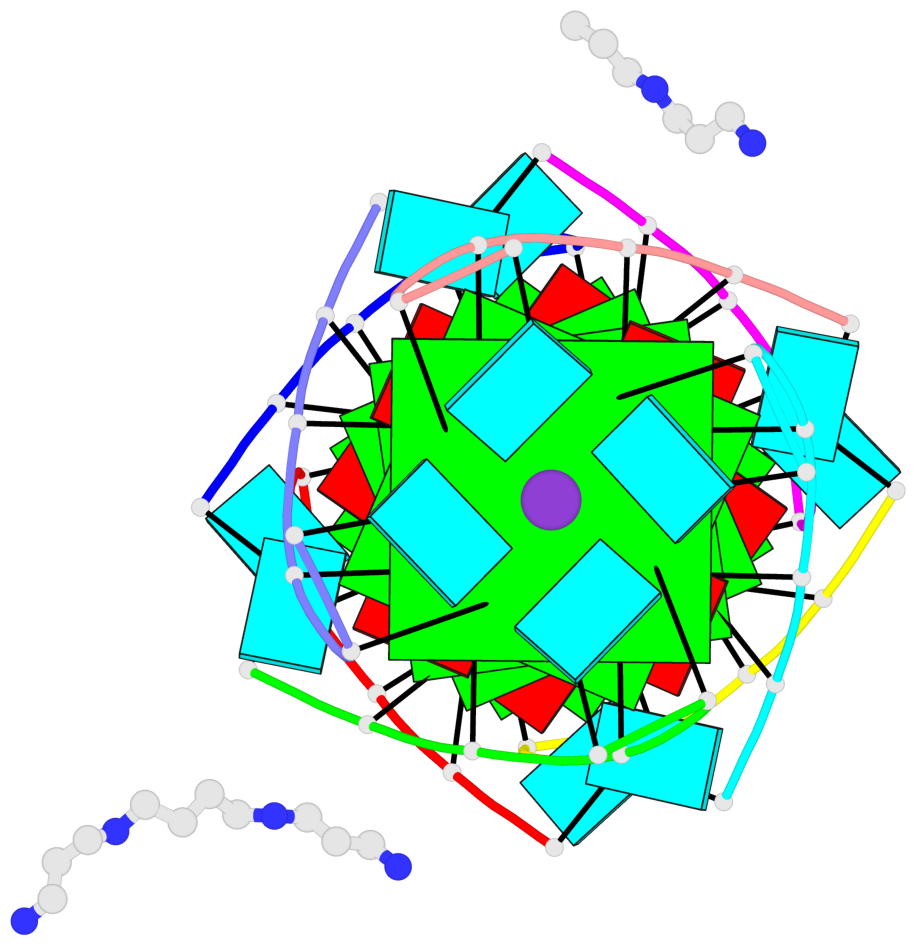
- 6 G-tetrads, 1 G4 helix, 2 G4 stems, 1 G4 coaxial stack, parallel(4+0), UUUU, coaxial interfaces: 5'/5'-SEPARATED
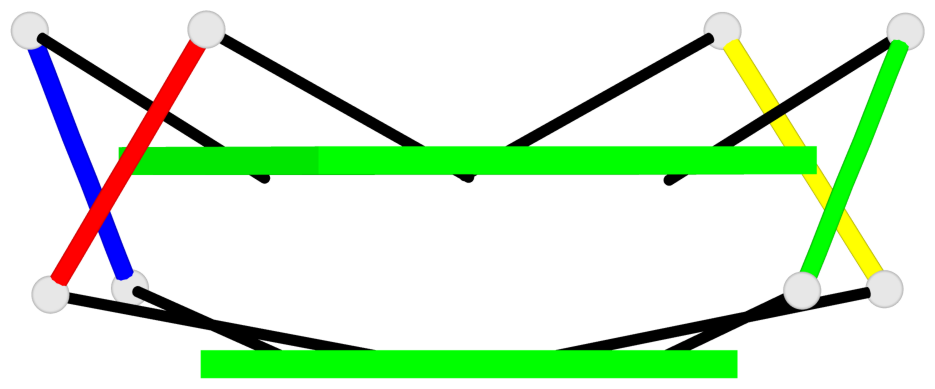
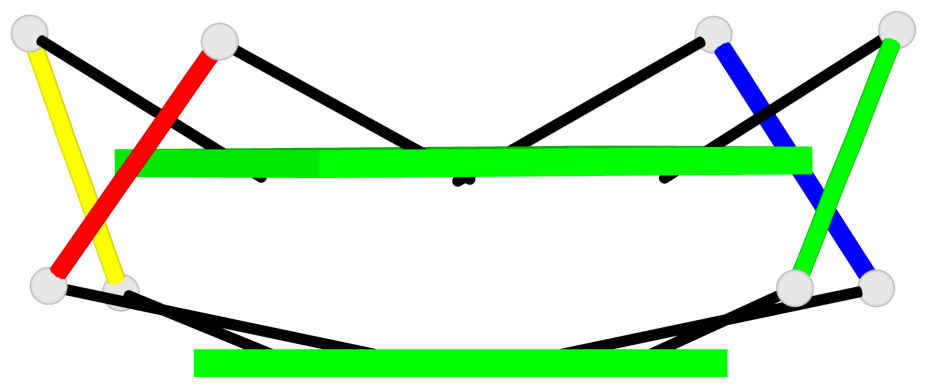
Base-block schematics in six views
List of 6 G-tetrads
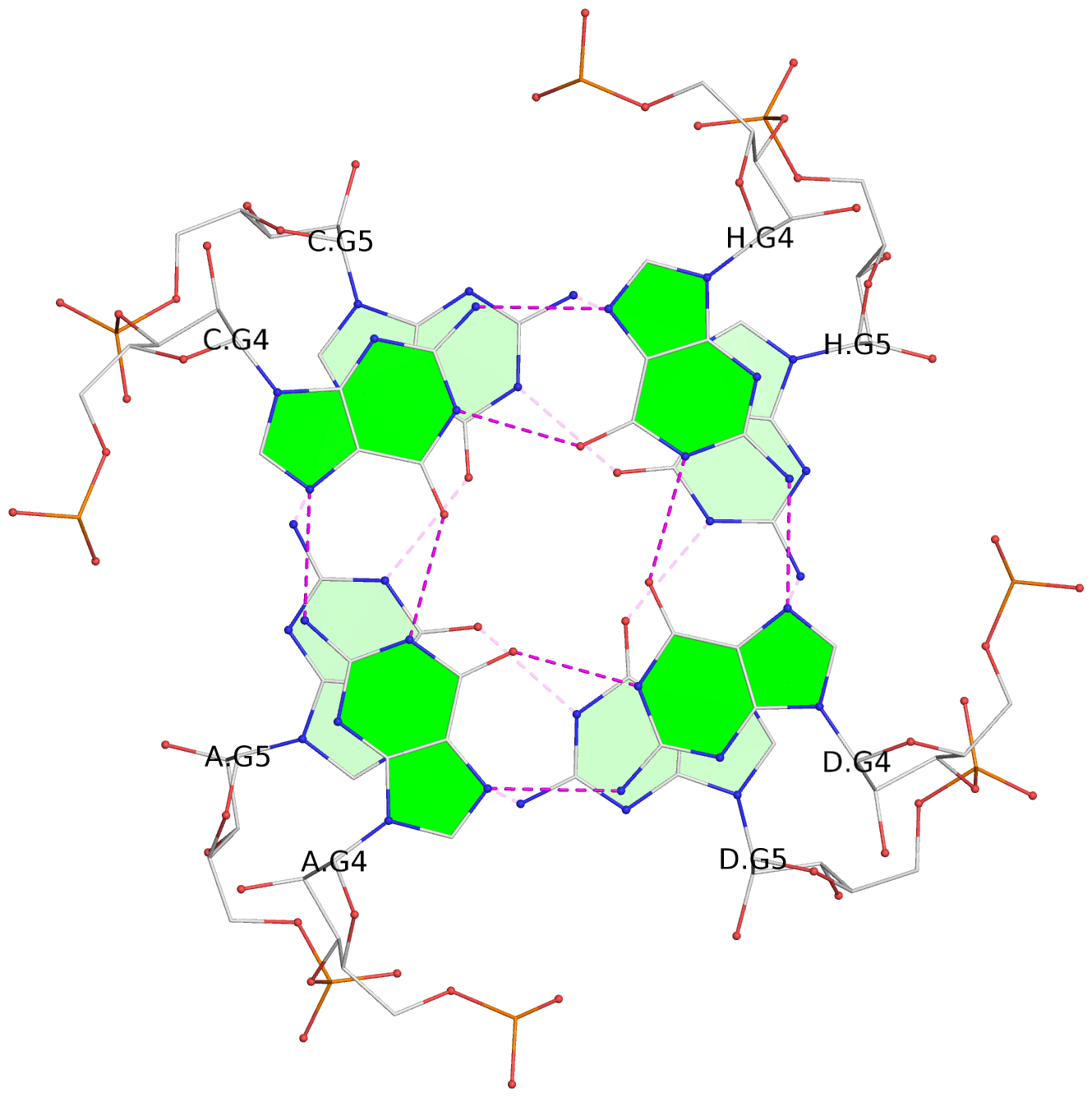
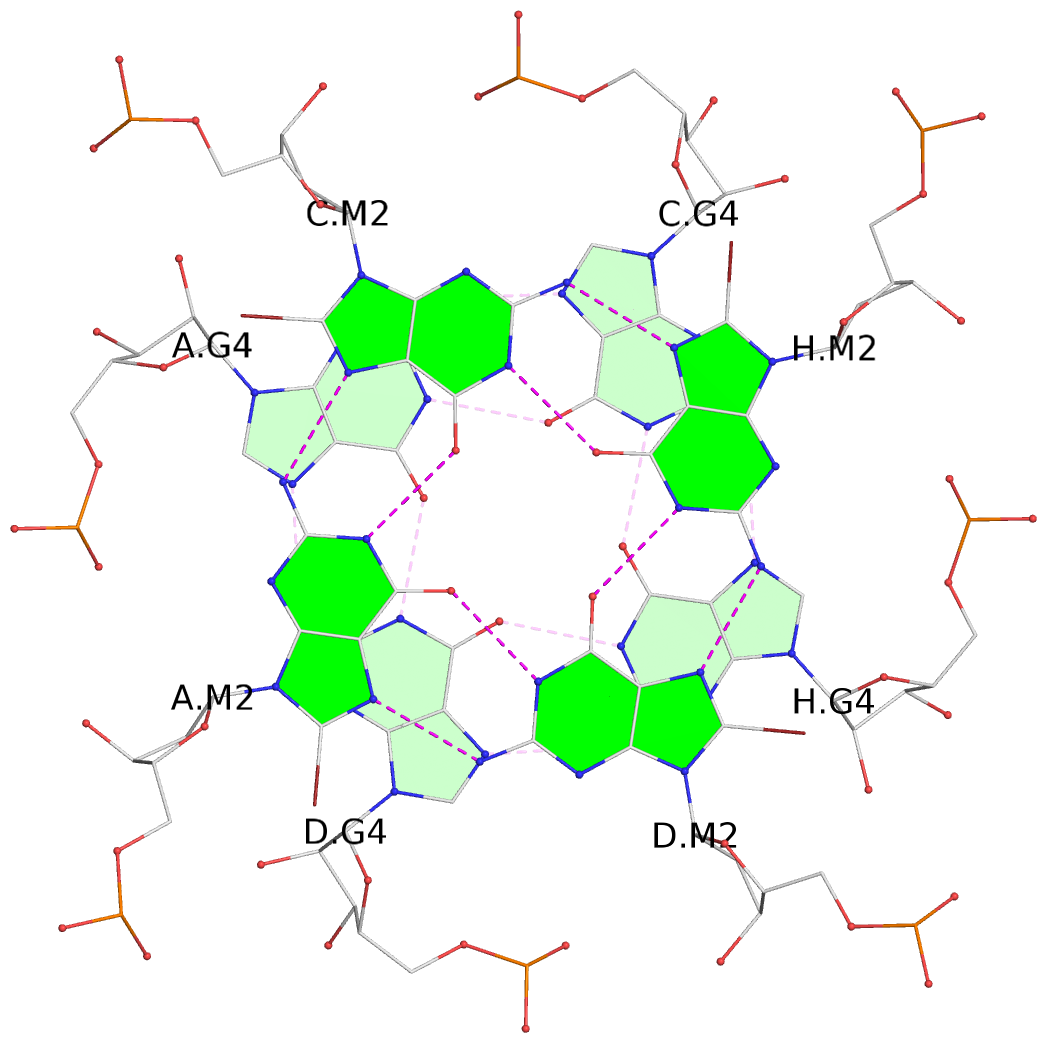
1 glyco-bond=---- sugar=---- groove=---- planarity=0.098 type=planar nts=4 gggg A.BGM2,C.BGM2,H.BGM2,D.BGM2 2 glyco-bond=---- sugar=3333 groove=---- planarity=0.151 type=planar nts=4 GGGG A.G4,C.G4,H.G4,D.G4 3 glyco-bond=---- sugar=3333 groove=---- planarity=0.264 type=bowl nts=4 GGGG A.G5,C.G5,H.G5,D.G5 4 glyco-bond=---- sugar=---- groove=---- planarity=0.082 type=planar nts=4 gggg B.BGM2,E.BGM2,F.BGM2,G.BGM2 5 glyco-bond=---- sugar=3333 groove=---- planarity=0.154 type=planar nts=4 GGGG B.G4,E.G4,F.G4,G.G4 6 glyco-bond=---- sugar=3333 groove=---- planarity=0.257 type=bowl nts=4 GGGG B.G5,E.G5,F.G5,G.G5
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 6 G-tetrad layers, inter-molecular, with 2 stems
List of 2 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 2 G-tetrad layers, 0 loops, inter-molecular, UUUU, parallel, parallel(4+0)
Stem#2, 2 G-tetrad layers, 0 loops, inter-molecular, UUUU, parallel, parallel(4+0)
List of 1 G4 coaxial stack
1 G4 helix#1 contains 2 G4 stems: [#1,#2] [5'/5'-SEPARATED]