Detailed DSSR results for the G-quadruplex: PDB entry 1np9
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 1np9
- Class
- DNA
- Method
- NMR
- Summary
- Structure of the parallel-stranded DNA quadruplex d(ttaggga)4 containing the human telomeric repeat
- Reference
- Gavathiotis E, Searle MS (2003): "Structure of the parallel-stranded DNA quadruplex d(TTAGGGT)4 containing the human telomeric repeat: evidence for A-tetrad formation from NMR and molecular dynamics simulations." ORG.BIOMOL.CHEM., 1, 1650-1656. doi: 10.1039/b300845m.
- Abstract
- The structure of the intermolecular DNA quadruplex d(TTAGGGT)4, based on the human telomeric DNA sequence d(TTAGGG), has been determined in solution by NMR and restrained molecular dynamics simultations. The core GGG region forms a highly stable quadruplex with G-tetrads likely stabilised by K+ ions bound between tetrad plains. However, we have focused on the conformation of the adenines which differ considerably in base alignment, stability and dynamics from those in previously reported structures of d(AGGGT)4 and d(TAGGGT)4. We show unambiguously that the adenines of d(TTAGGGT)4 are involved in the formation of a relatively stable A-tetrad with well-defined glycosidic torsion angles (anti), hydrogen bonding network (adenine 6-NH2-adenine N1) defined by interbase NOEs, and base stacking interactions with the neighbouring G-tetrad. All of these structural features are apparent from NOE data involving both exchangeable and non-exchangeable protons. Thus, context-dependent effects appear to play some role in dictating preferred conformation, stability and dynamics. The structure of d(TTAGGGT)4 provides us with a model system for exploiting in the design of novel telomerase inhibitors that bind to and stabilise G-quadruplex structures.
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, parallel(4+0), UUUU
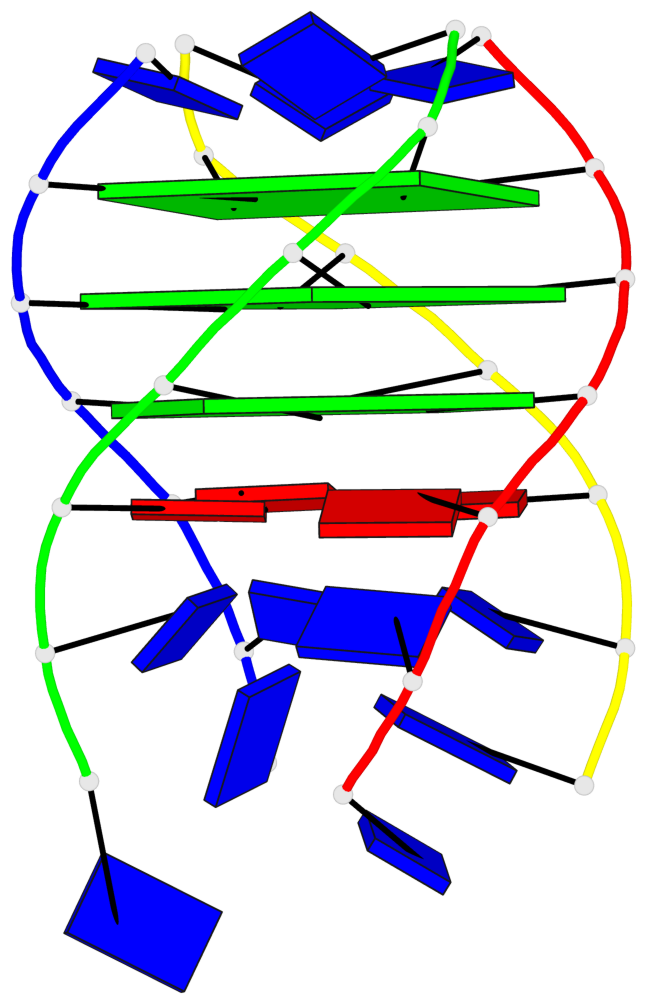
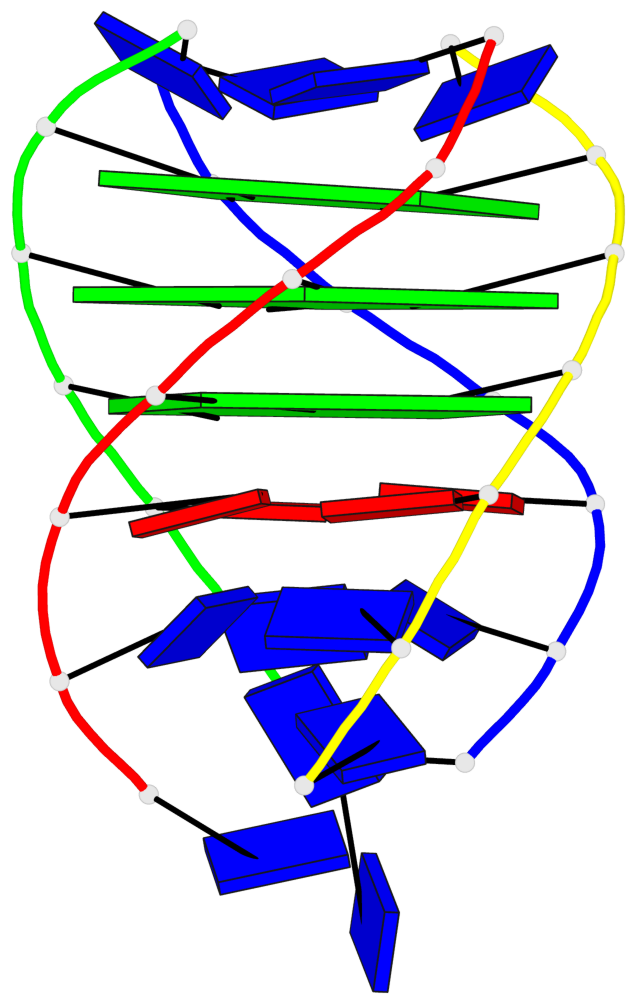
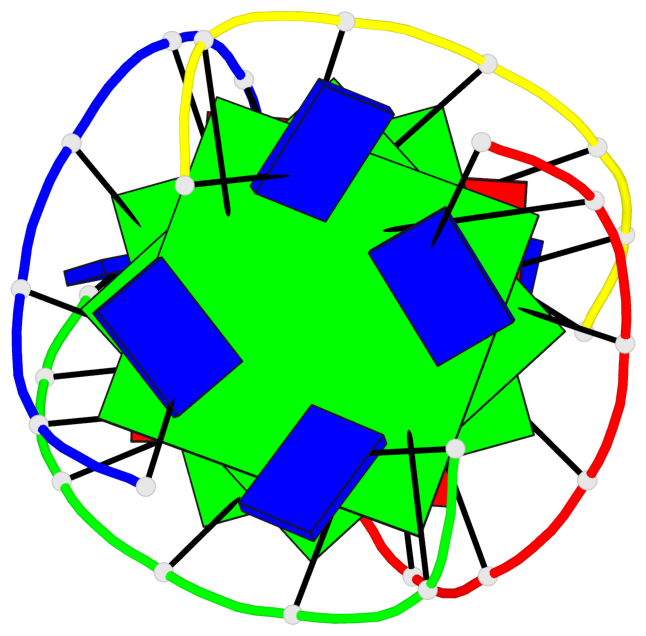
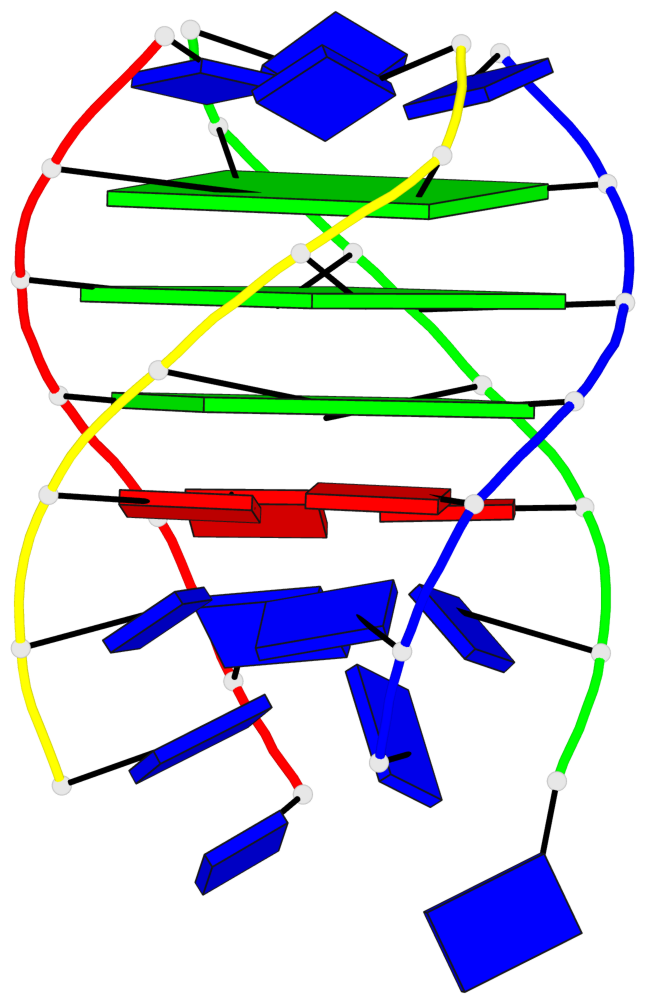
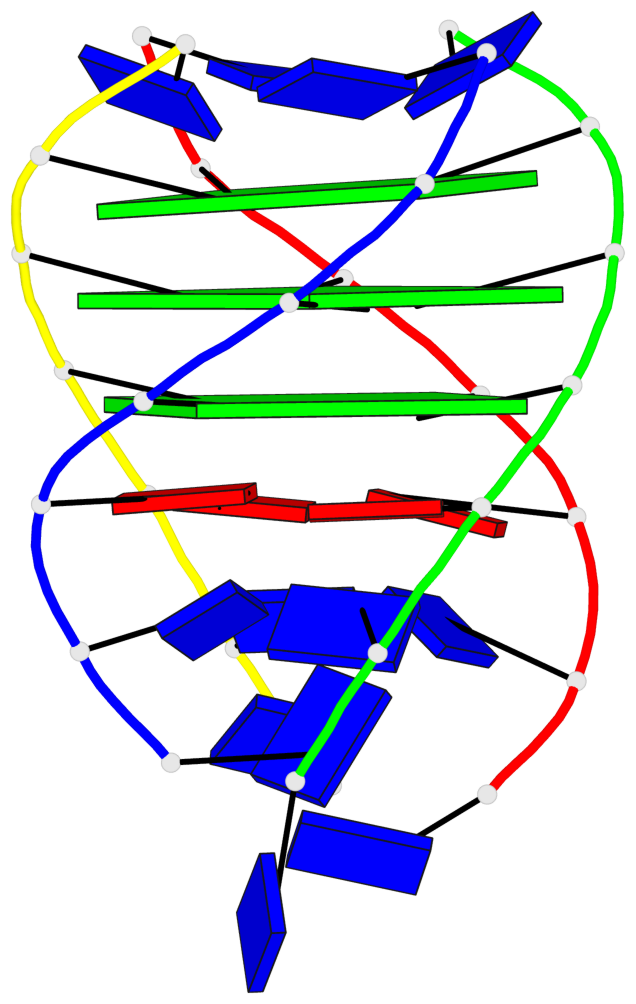
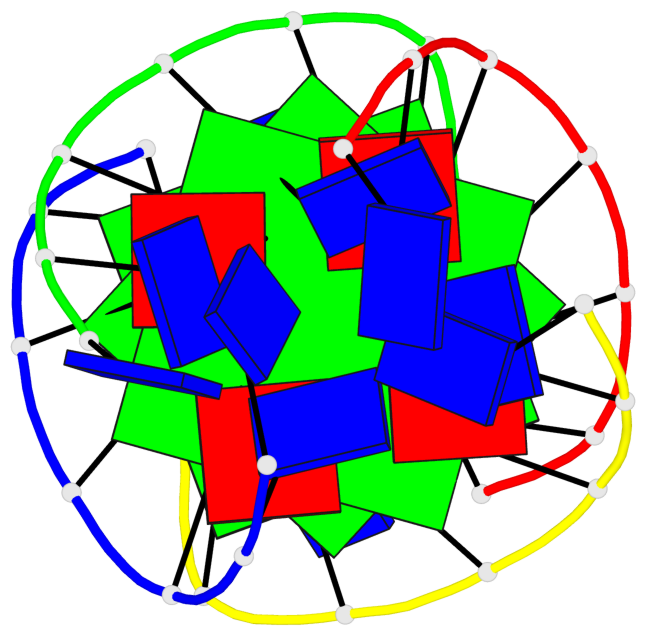
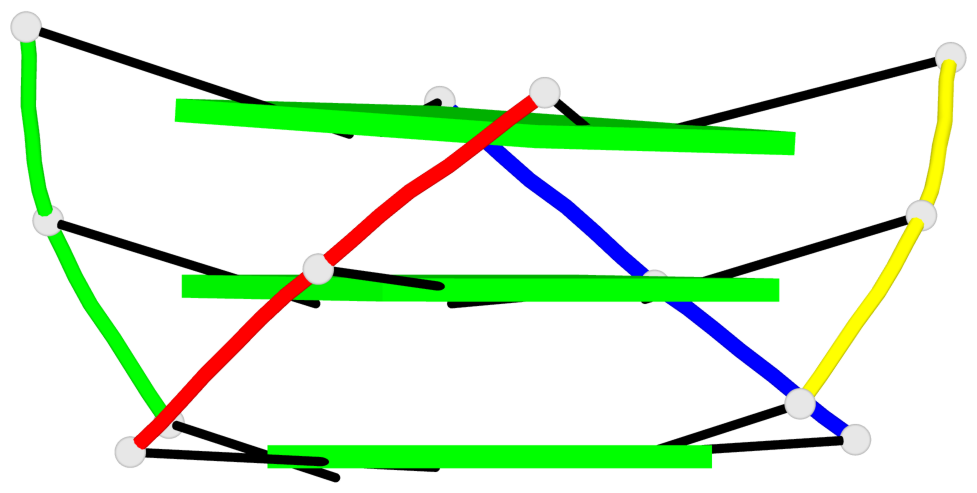
Base-block schematics in six views
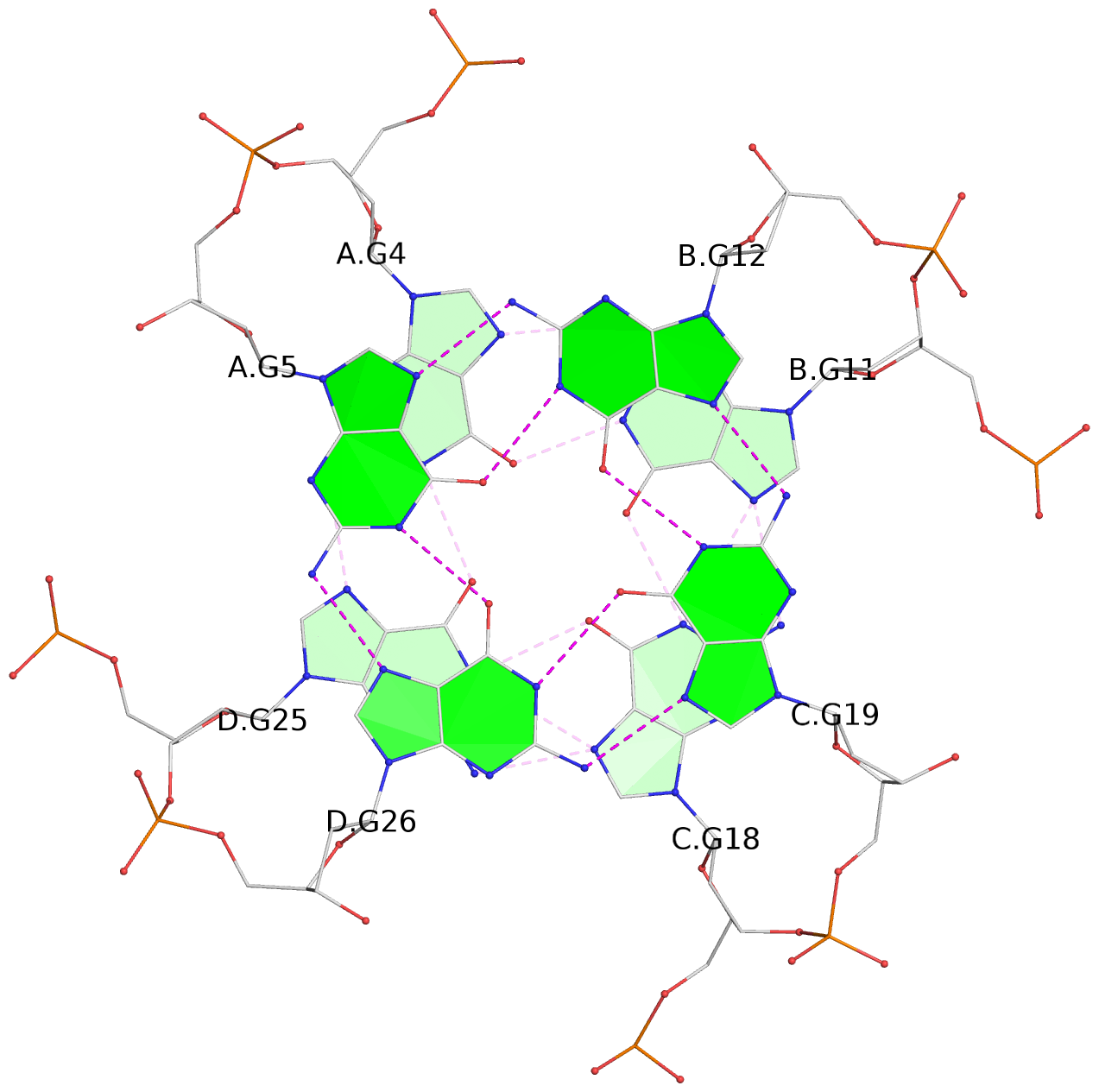
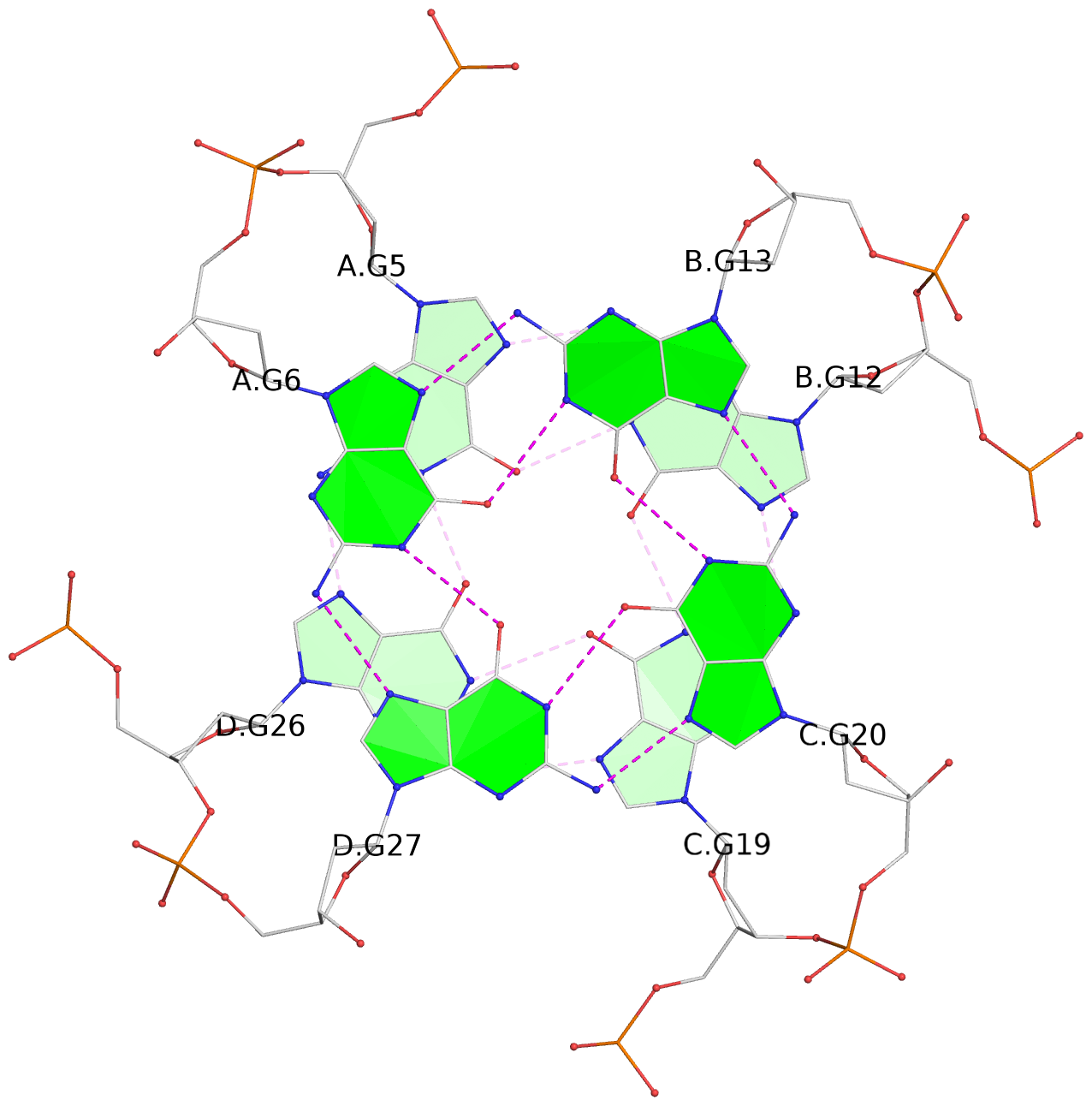
List of 3 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.219 type=other nts=4 GGGG A.DG4,D.DG25,C.DG18,B.DG11 2 glyco-bond=---- sugar=---- groove=---- planarity=0.271 type=bowl nts=4 GGGG A.DG5,D.DG26,C.DG19,B.DG12 3 glyco-bond=---- sugar=---- groove=---- planarity=0.282 type=other nts=4 GGGG A.DG6,D.DG27,C.DG20,B.DG13
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, inter-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.