Detailed DSSR results for the G-quadruplex: PDB entry 1nyd
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 1nyd
- Class
- DNA
- Method
- NMR
- Summary
- Solution structure of DNA quadruplex gcggtggat
- Reference
- Webba da Silva M (2003): "Association of DNA quadruplexes through G:C:G:C tetrads. Solution structure of d(GCGGTGGAT)." Biochemistry, 42, 14356-14365. doi: 10.1021/bi0355185.
- Abstract
- The structure formed by the DNA sequence d(GCGGTGGAT) in a 100 mM Na(+) solution has been determined using molecular dynamics calculations constrained by distance and dihedral restraints derived from NMR experiments performed at isotopic natural abundance. The sequence folds into a dimer of dimers. Each symmetry-related half contains two parallel stranded G:G:G:G tetrads flanked by an A:A mismatch and by four-stranded G:C:G:C tetrads. Each of the two juxtaposed G:C:G:C tetrads is composed of alternating antiparallel strands from the two halves of the dimer. For each single strand, a thymine intersperses a double chain reversal connecting the juxtaposed G:G:G:G tetrads. This architecture has potential implications in genetic recombination. It suggests a pathway for oligomerization involving association of quadruplex entities through GpC steps.
- G4 notes
- 4 G-tetrads, 2 G4 helices, 2 G4 stems, parallel(4+0), UUUU
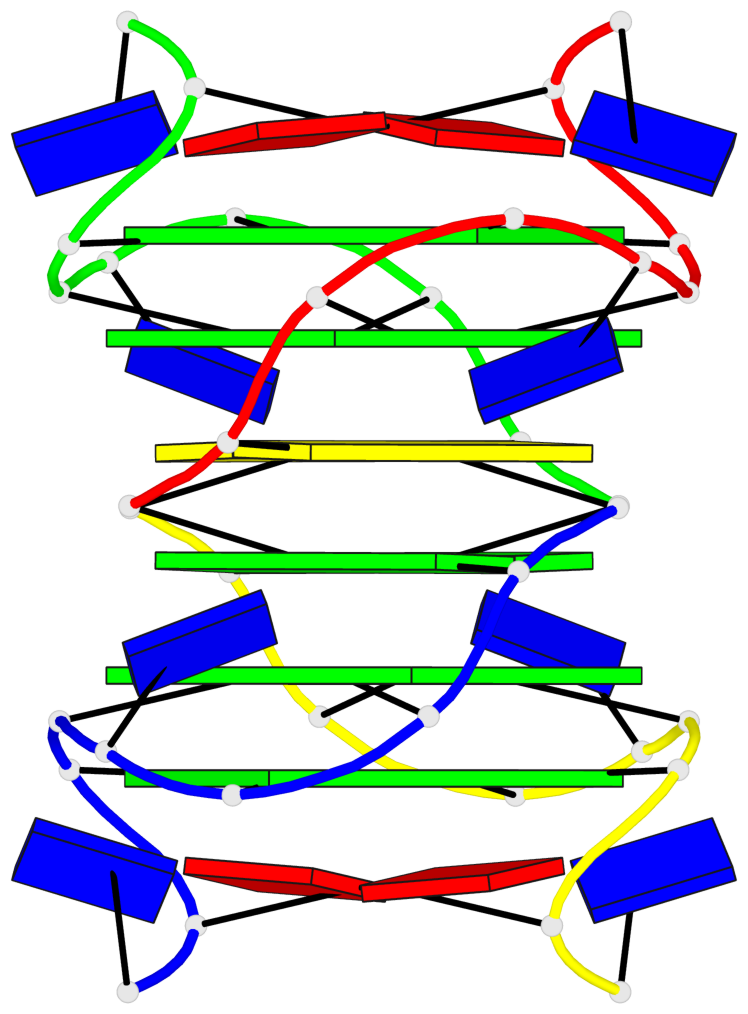
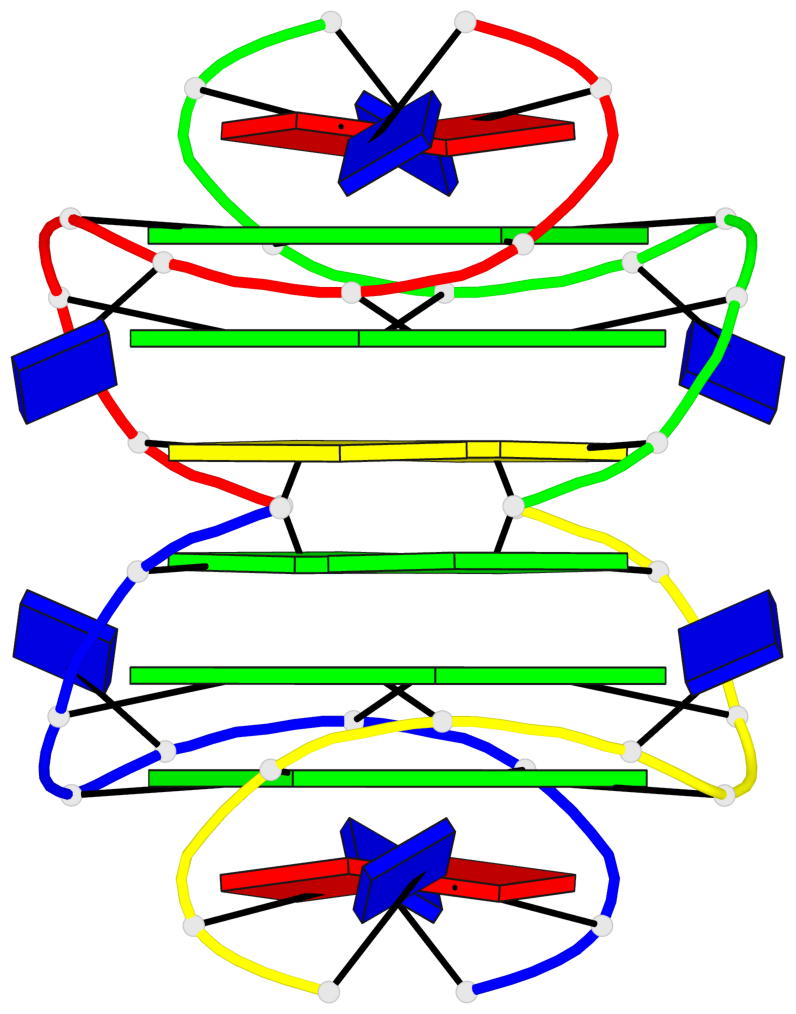
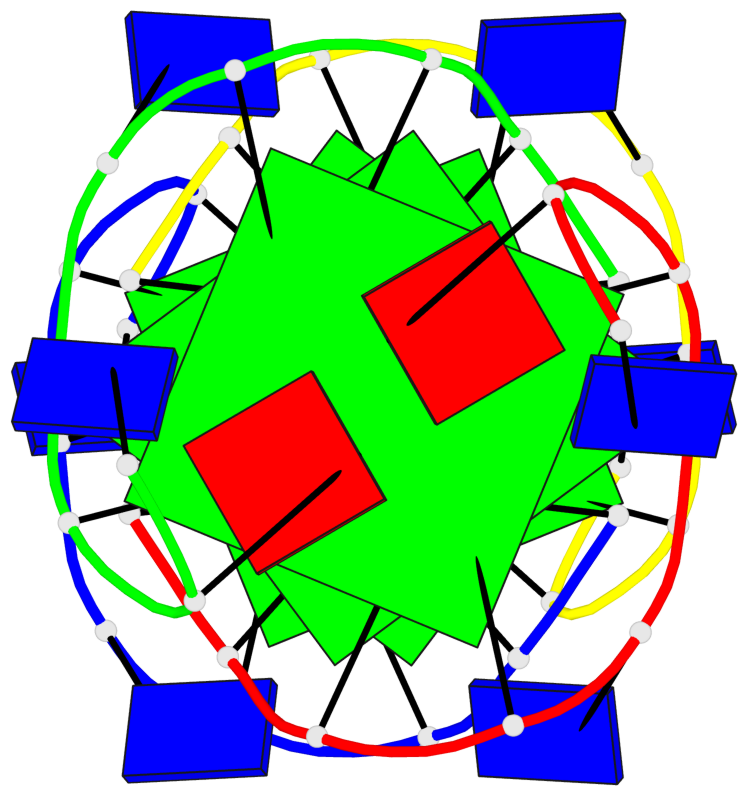
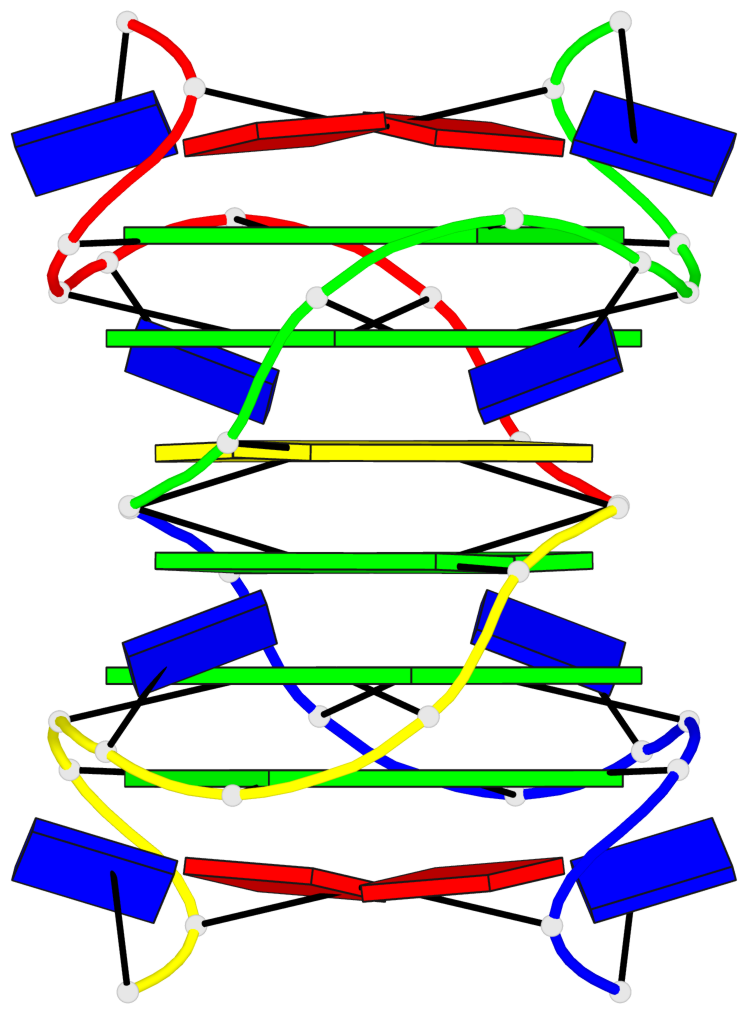
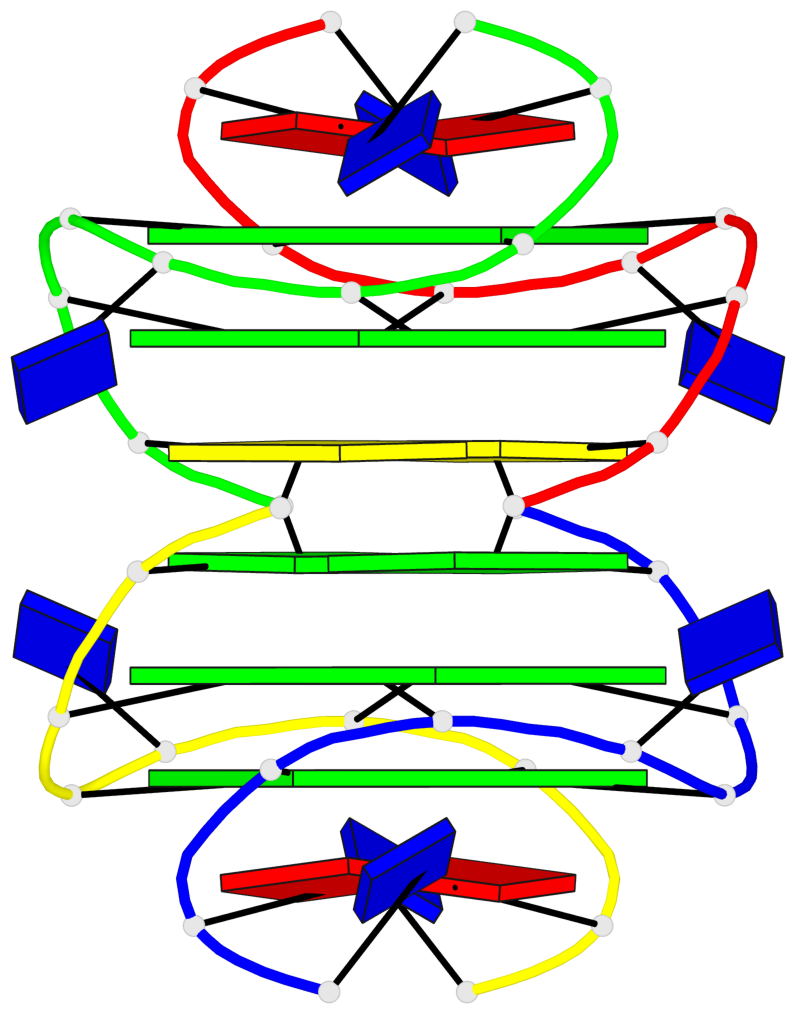
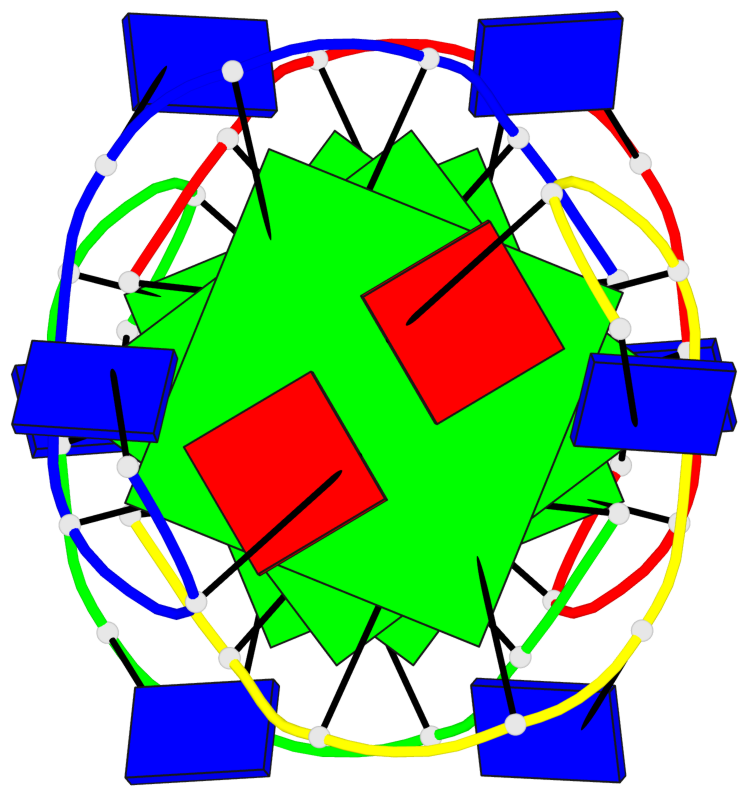
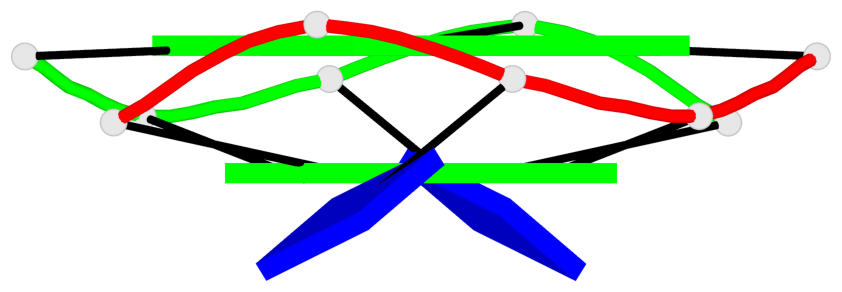
Base-block schematics in six views
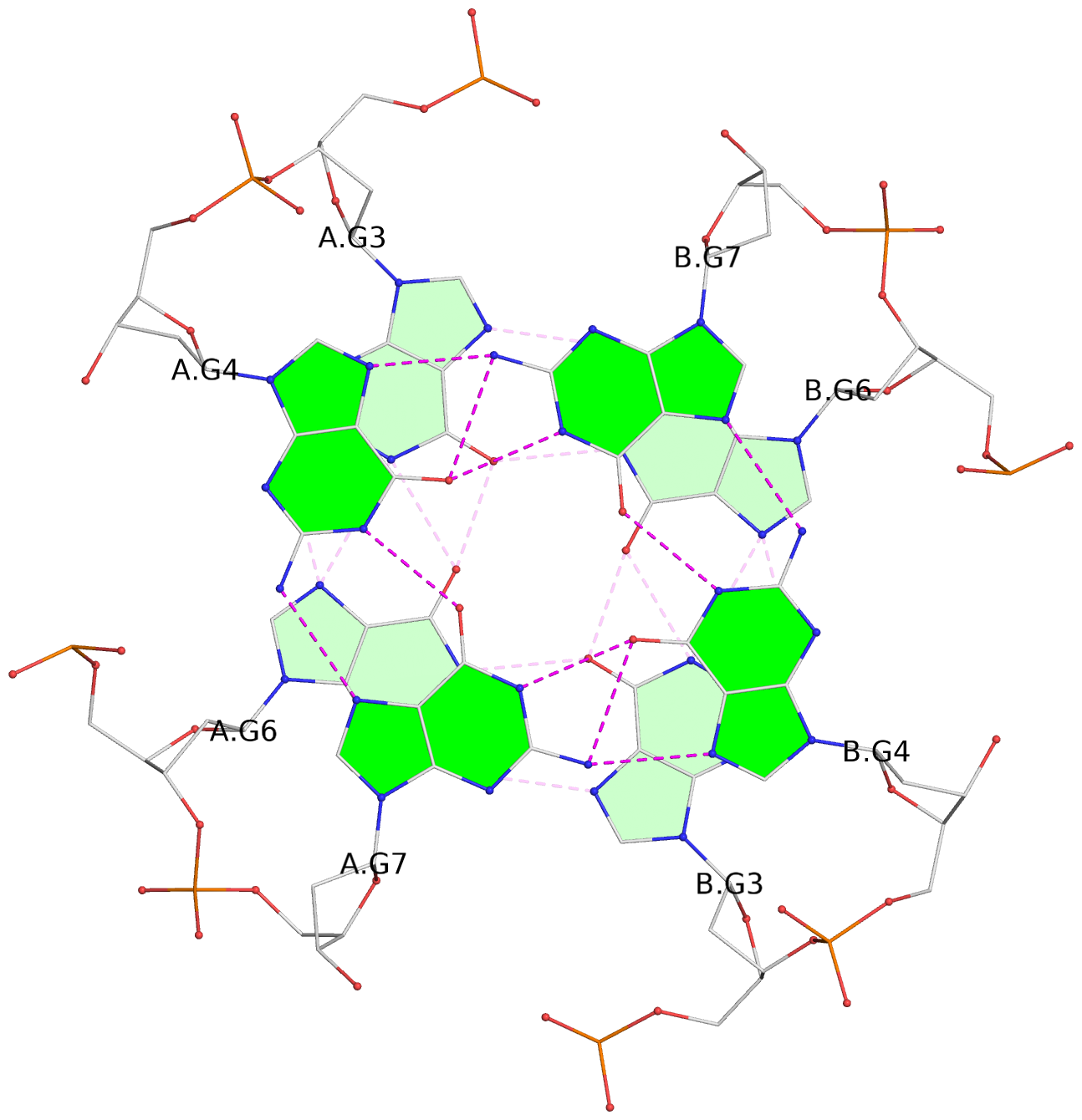
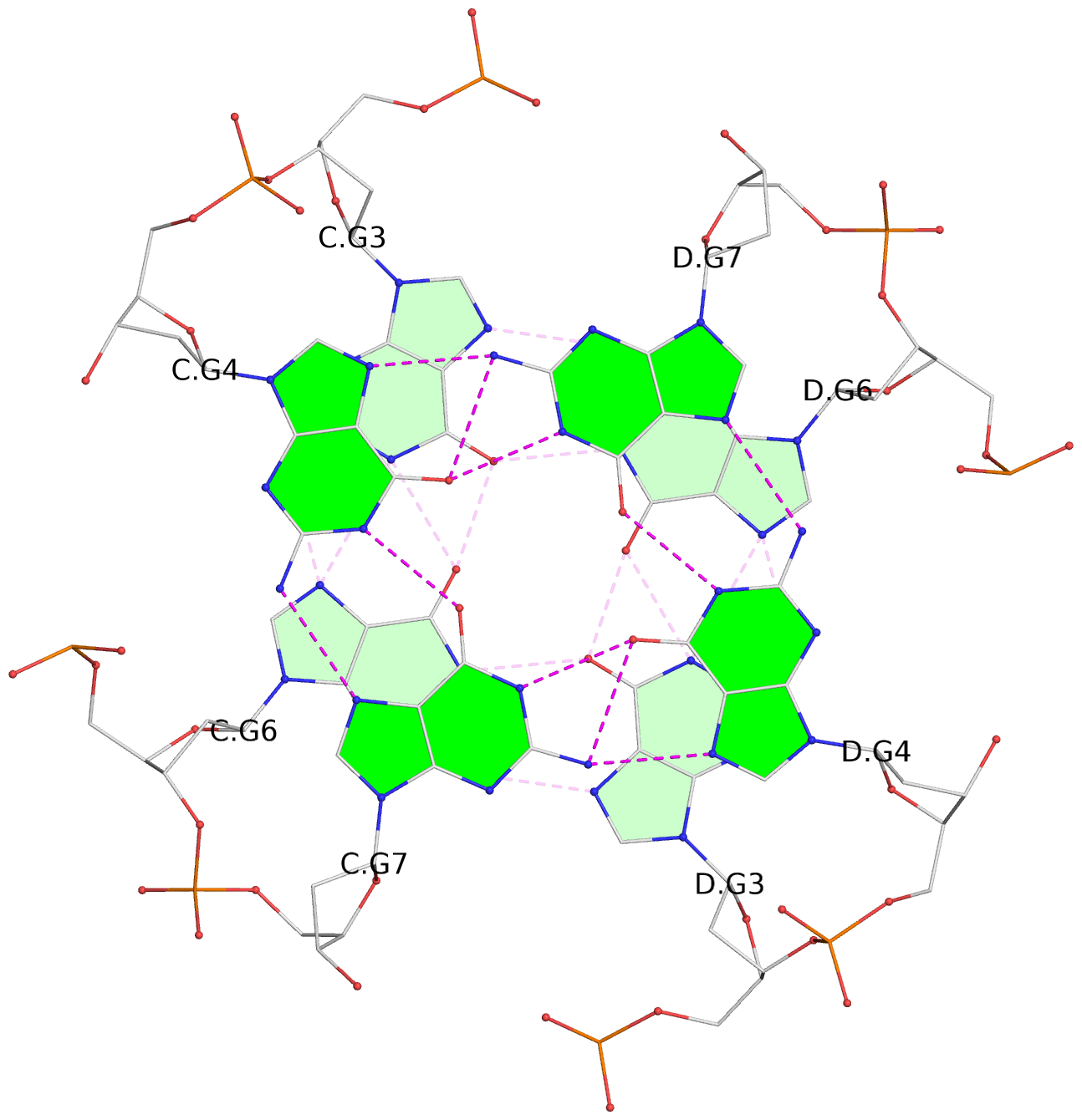
List of 4 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.139 type=planar nts=4 GGGG A.DG3,A.DG6,B.DG3,B.DG6 2 glyco-bond=---- sugar=---- groove=---- planarity=0.053 type=planar nts=4 GGGG A.DG4,A.DG7,B.DG4,B.DG7 3 glyco-bond=---- sugar=---- groove=---- planarity=0.139 type=planar nts=4 GGGG C.DG3,C.DG6,D.DG3,D.DG6 4 glyco-bond=---- sugar=---- groove=---- planarity=0.053 type=planar nts=4 GGGG C.DG4,C.DG7,D.DG4,D.DG7
List of 2 G4-helices
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 2 G-tetrad layers, inter-molecular, with 1 stem
Helix#2, 2 G-tetrad layers, inter-molecular, with 1 stem
List of 2 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.