Detailed DSSR results for the G-quadruplex: PDB entry 1nzm
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 1nzm
- Class
- DNA
- Method
- NMR
- Summary
- NMR structure of the parallel-stranded DNA quadruplex d(ttagggt)4 complexed with the telomerase inhibitor rhps4
- Reference
- Gavathiotis E, Heald RA, Stevens MFG, Searle MS (2003): "Drug Recognition and Stabilisation of the Parallel-stranded DNA Quadruplex d(TTAGGGT)4 Containing the Human Telomeric Repeat." J.Mol.Biol., 334, 25-36. doi: 10.1016/j.jmb.2003.09.018.
- Abstract
- The NMR structure of the parallel-stranded DNA quadruplex d(TTAGGGT)(4), containing the human telomeric repeat, has been determined in solution in complex with a fluorinated pentacyclic quino[4,3,2-kl]acridinium cation (RHPS4). RHPS4 has been identified as a potent inhibitor of telomerase at submicromolar levels (IC(50) value of 0.33(+/-0.13)microM), exhibiting a wide differential between telomerase inhibition and acute cellular toxicity. All of the data point to RHPS4 exerting its chemotherapeutic potency through interaction with, and stabilisation of, four-stranded G-quadruplex structures. RHPS4 forms a dynamic interaction with d(TTAGGGT)(4), as evident from 1H and 19F linewidths, with fast exchange between binding sites induced at 318 K. Perturbations to DNA chemical shifts and 24 intermolecular nuclear Overhauser effects (NOEs) identify the 5'-ApG and 5'-GpT steps as the principle intercalation sites; a structural model has been refined using NOE-restrained molecular dynamics. The central G-tetrad core remains intact, with drug molecules stacking at the ends of the G-quadruplex. The partial positive charge on position 13-N of the acridine ring appears to act as a "pseudo" potassium ion and is positioned above the centre of the G-tetrad in the region of high negative charge density. In both ApG and GpT intercalation sites, the drug is seen to converge to the same orientation in which the pi-system of the drug overlaps primarily with two bases of each G-tetrad. The drug is held in place by stacking interactions with the G-tetrads; however, there is some evidence for a more dynamic, weakly stabilised A-tetrad that stacks partially on top of the drug at the 5'-end of the sequence. Together, the interactions of RHPS4 increase the t(m) of the quadruplex by approximately 20 degrees C. There is no evidence for drug intercalation within the G-quadruplex; however, the structural model strongly supports end-stacking interactions with the terminal G-tetrads.
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, parallel(4+0), UUUU
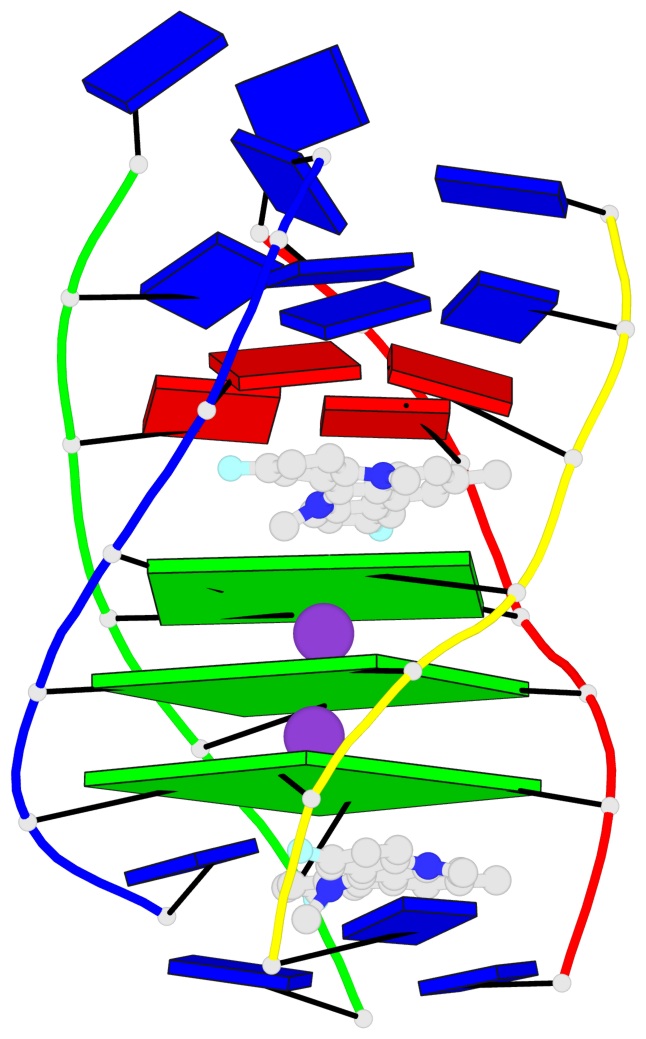
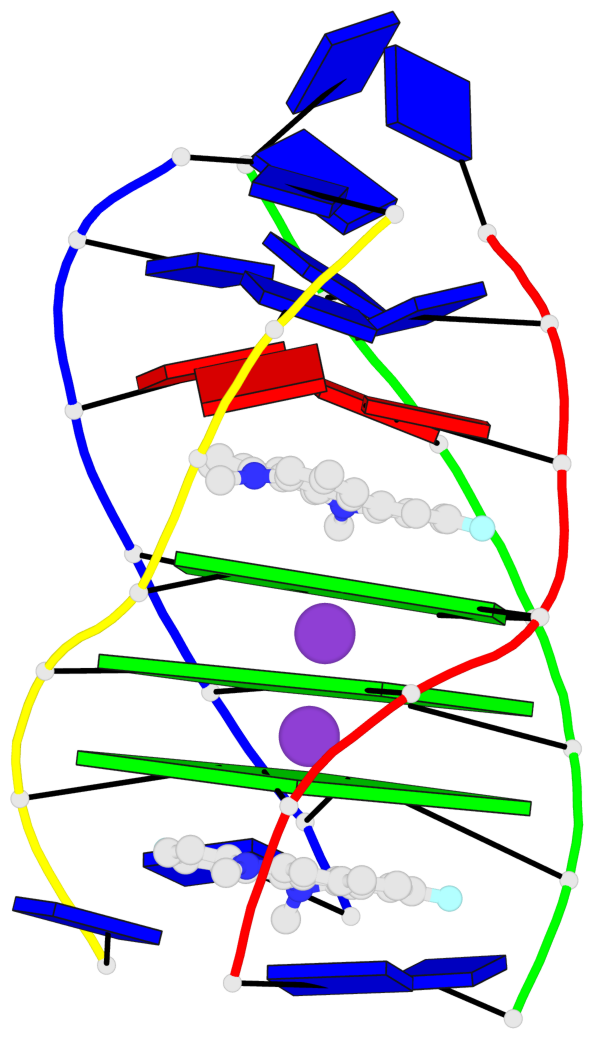
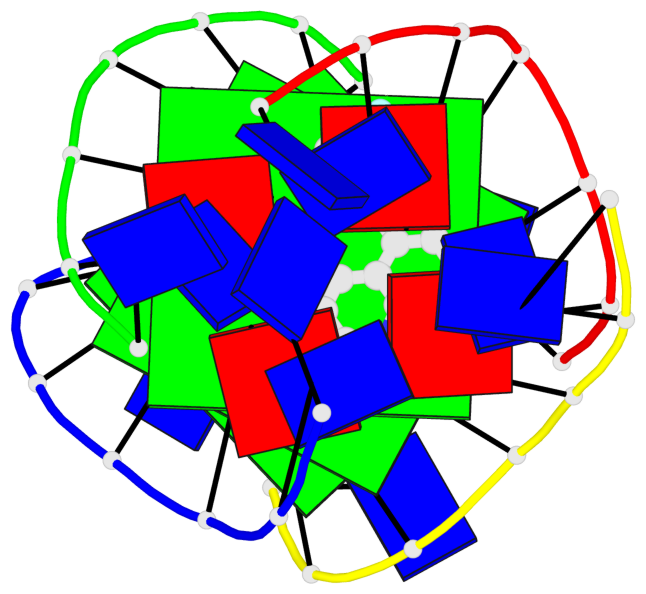
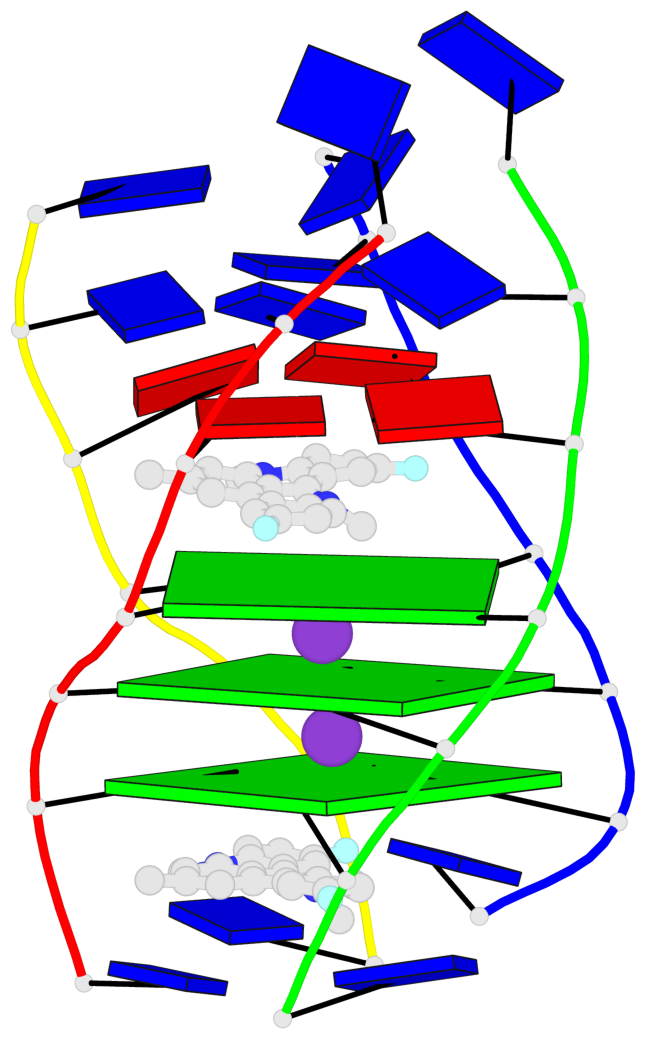
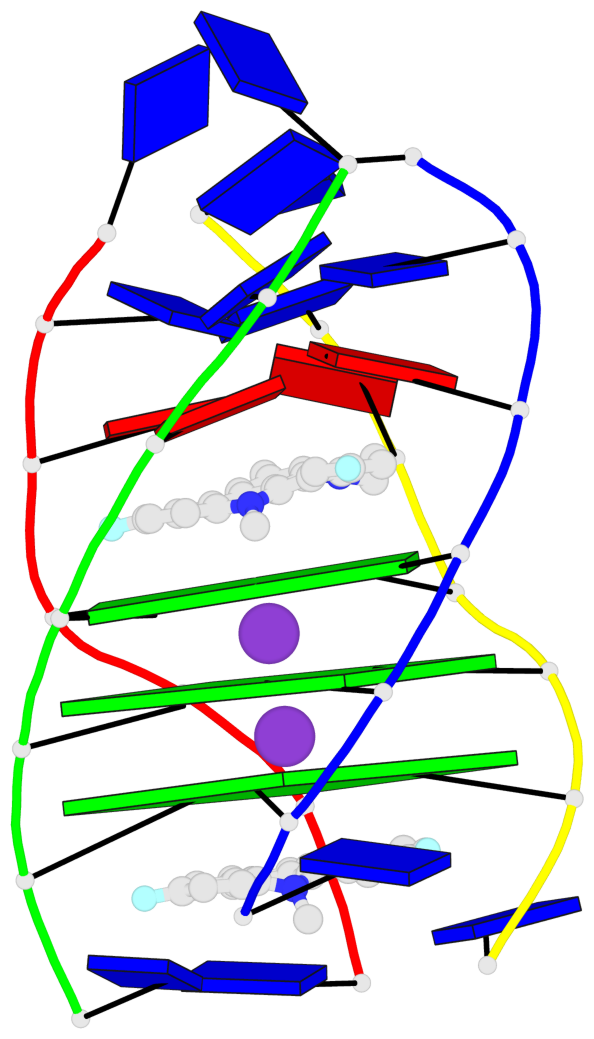
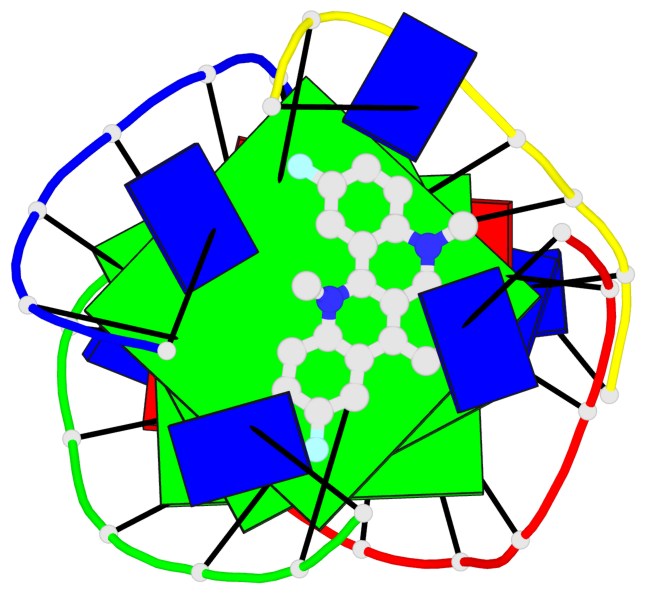
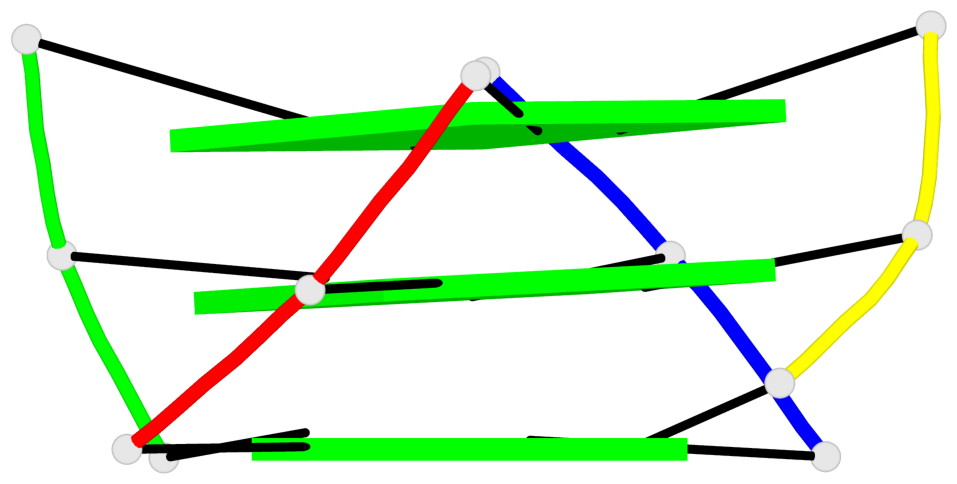
Base-block schematics in six views
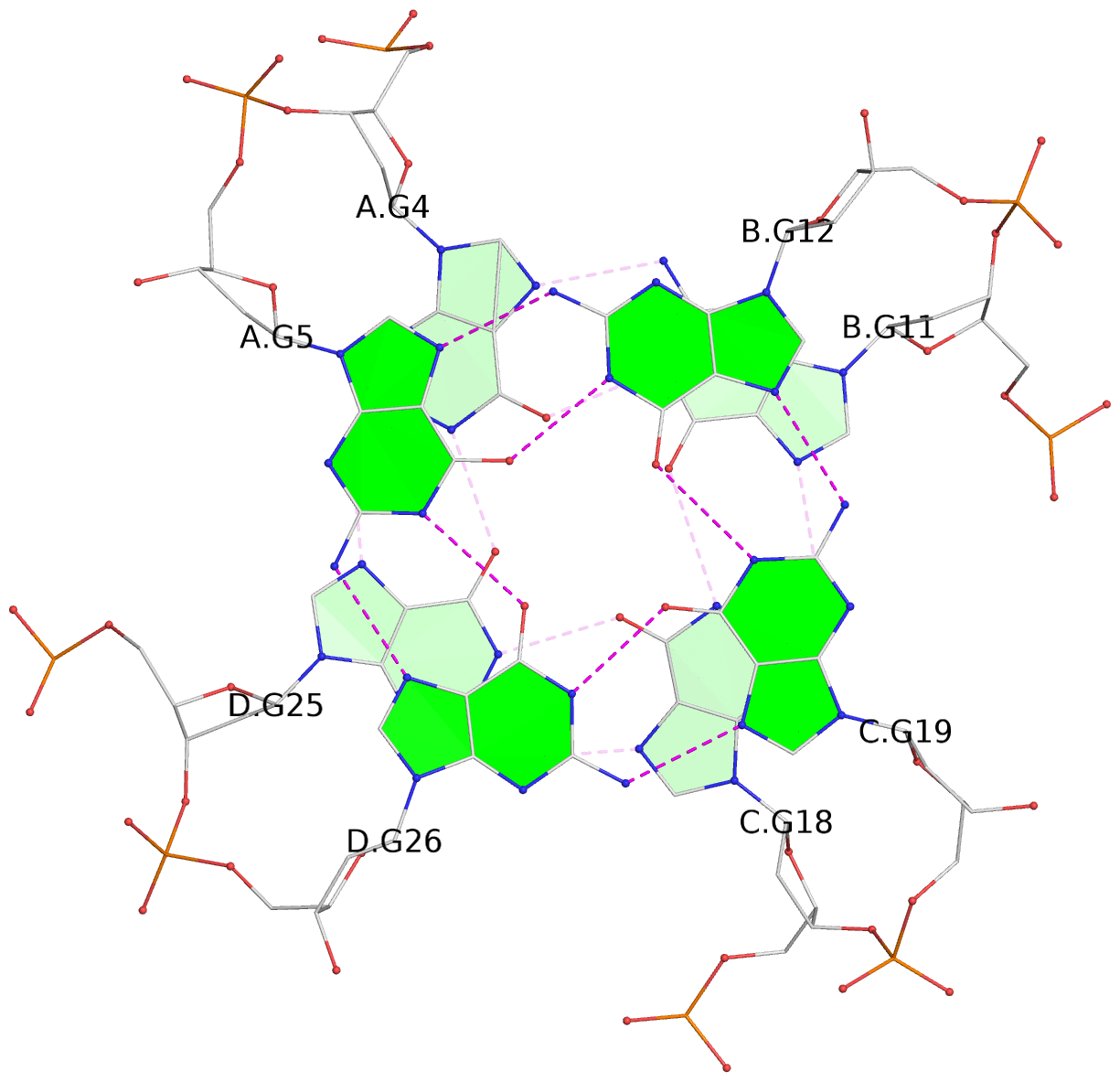
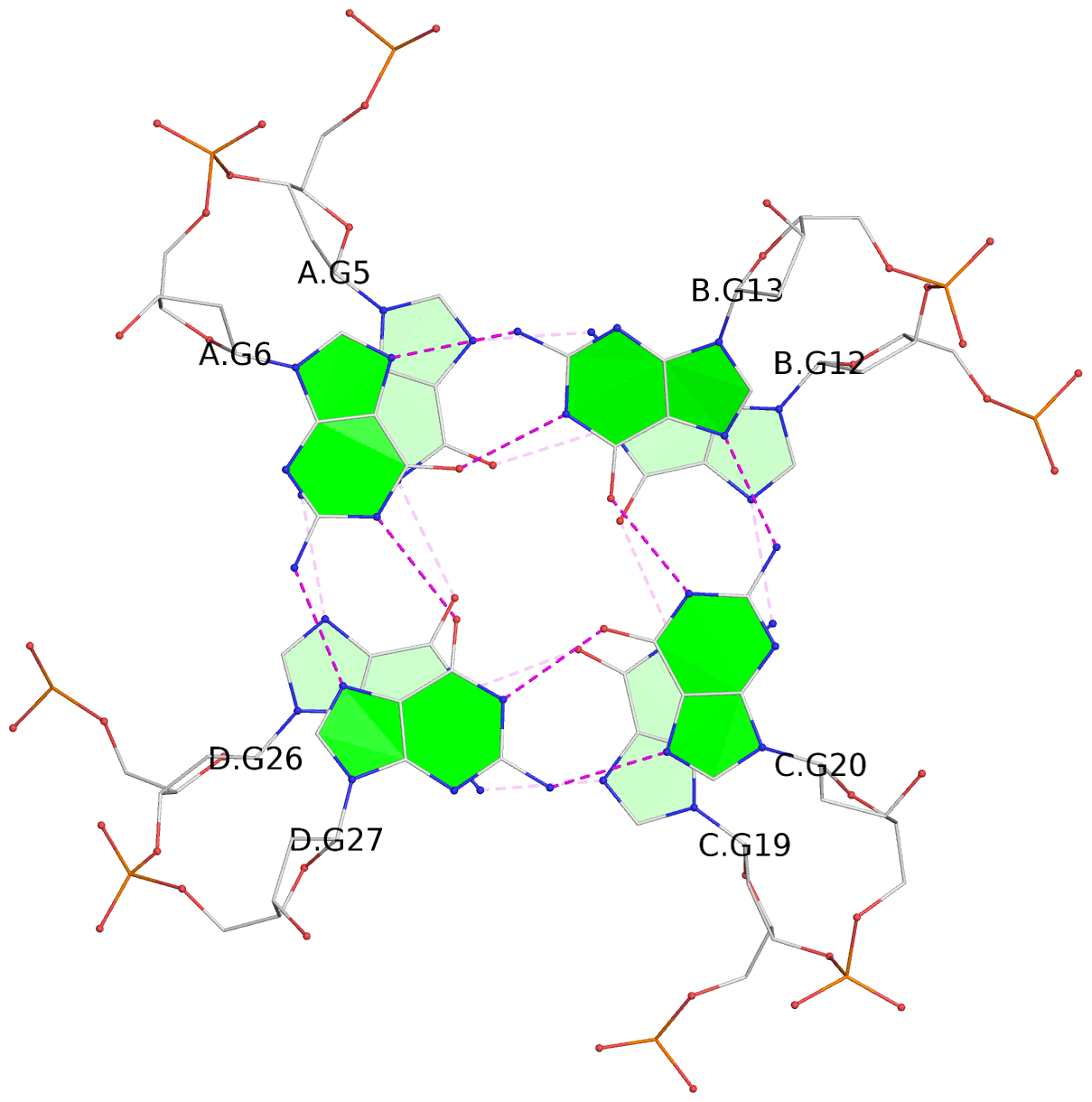
List of 3 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.345 type=other nts=4 GGGG A.DG4,D.DG25,C.DG18,B.DG11 2 glyco-bond=---- sugar=.-.- groove=---- planarity=0.264 type=other nts=4 GGGG A.DG5,D.DG26,C.DG19,B.DG12 3 glyco-bond=---- sugar=---- groove=---- planarity=0.317 type=other nts=4 GGGG A.DG6,D.DG27,C.DG20,B.DG13
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, inter-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.