Detailed DSSR results for the G-quadruplex: PDB entry 1oz8
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 1oz8
- Class
- DNA
- Method
- NMR
- Summary
- Intramolecular higher-order packing of parallel quadruplexes comprising a g:g:g:g tetrad and a g(:a):g(:a):g(:a):g heptad of gga triplet repeat DNA
- Reference
- Matsugami A, Okuizumi T, Uesugi S, Katahira M (2003): "Intramolecular Higher Order Packing of Parallel Quadruplexes Comprising a G:G:G:G Tetrad and a G(:A):G(:A):G(:A):G Heptad of GGA Triplet Repeat DNA." J.BIOL.CHEM., 278, 28147-28153. doi: 10.1074/jbc.M303694200.
- Abstract
- GGA triplet repeats are widely dispersed throughout eukaryotic genomes and are frequently located within biologically important regions such as gene regulatory regions and recombination hot spot sites. We determined the structure of d(GGA)4 (12-mer) under physiological conditions and founded the formation of an intramolecular parallel quadruplex for the first time. Later, a similar architecture to that of the intramolecular parallel quadruplex was found for a telomere DNA in the crystalline state. Here, we have determined the structure of d(GGA)8 (24-mer) under physiological conditions. Two intramolecular parallel quadruplexes comprising a G:G:G:G tetrad and a G(:A):G(:A):G(:A):G heptad are formed in d(GGA)8. These quadruplexes are packed in a tail-to-tail manner. This is the first demonstration of the intramolecular higher order packing of quadruplexes at atomic resolution. K+ ions, but not Na+ ones, are critically required for the formation of this unique structure. The elucidated structure suggests the mechanisms underlying the biological events related to the GGA triplet repeat. Furthermore, in the light of the structure, the mode of the higher order packing of the telomere DNA is discussed.
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 2(-P-P-P), parallel(4+0), UUUU
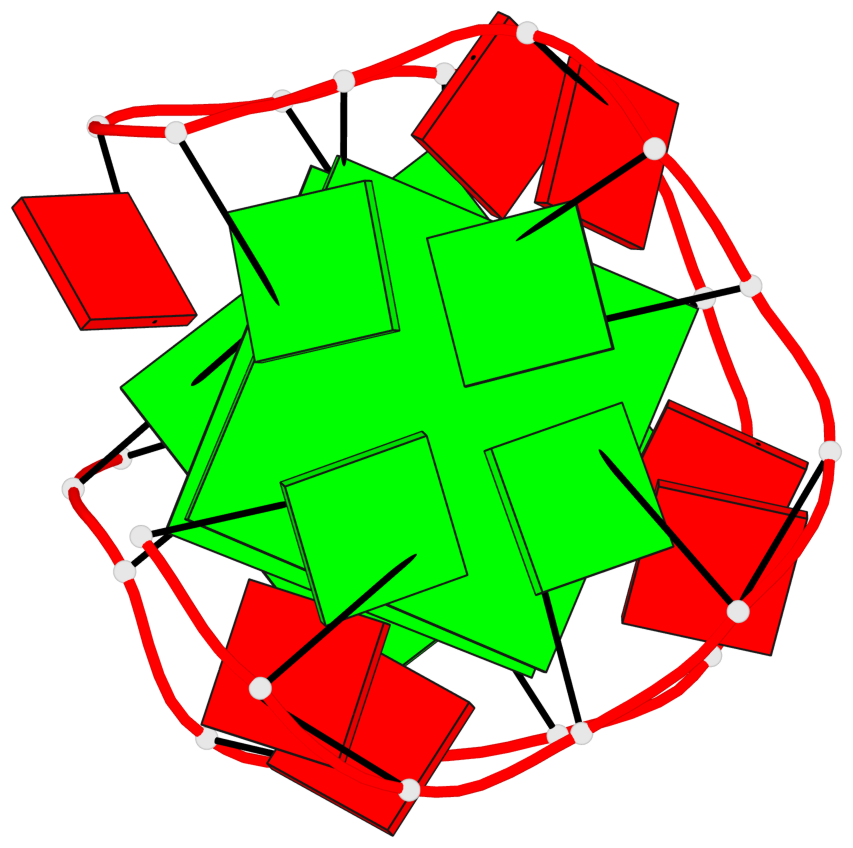
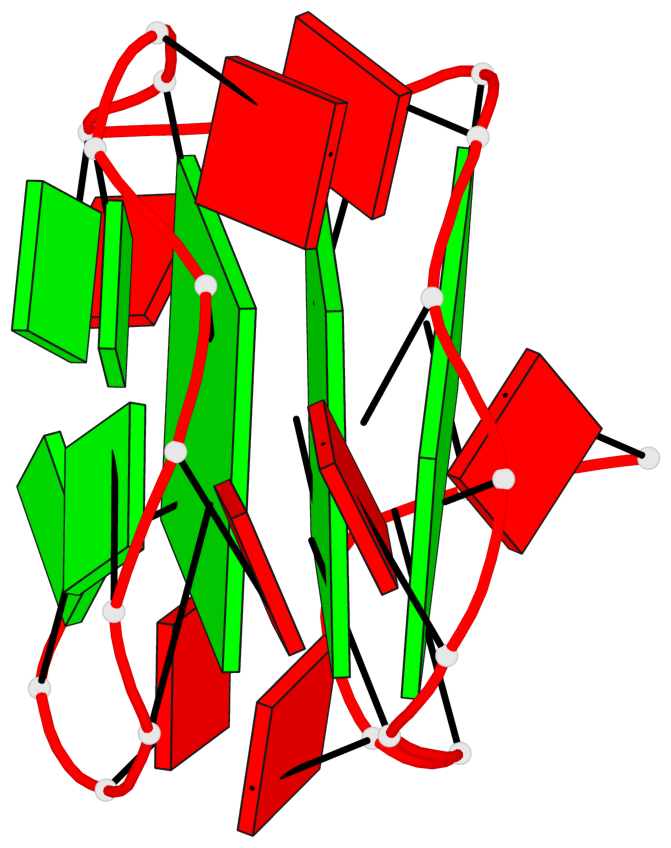
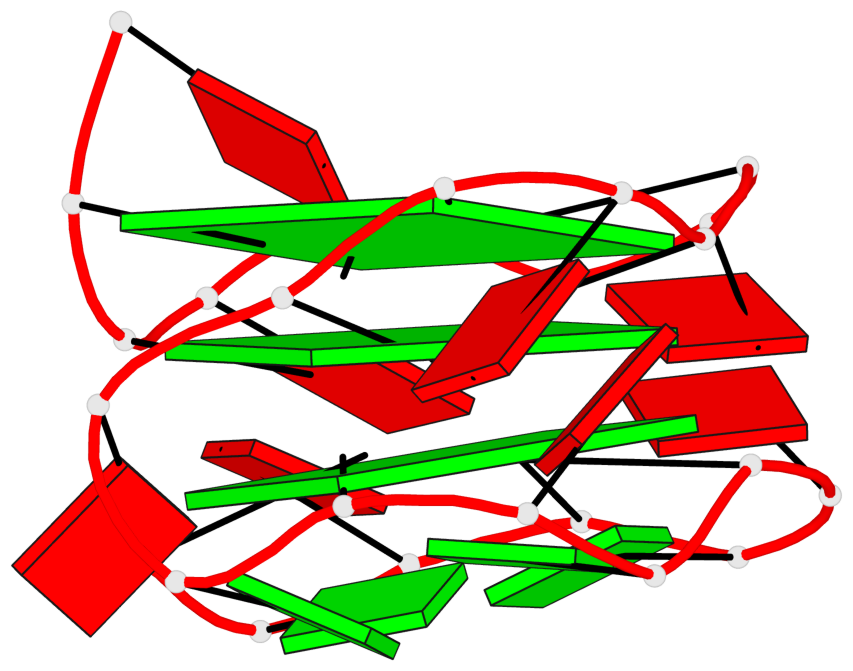
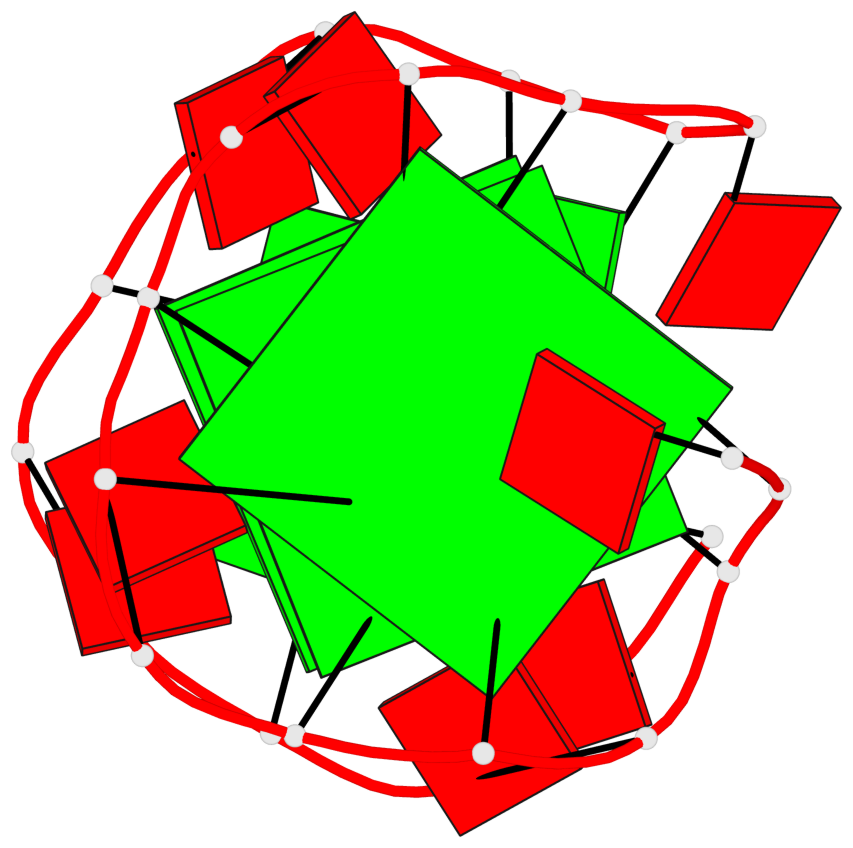
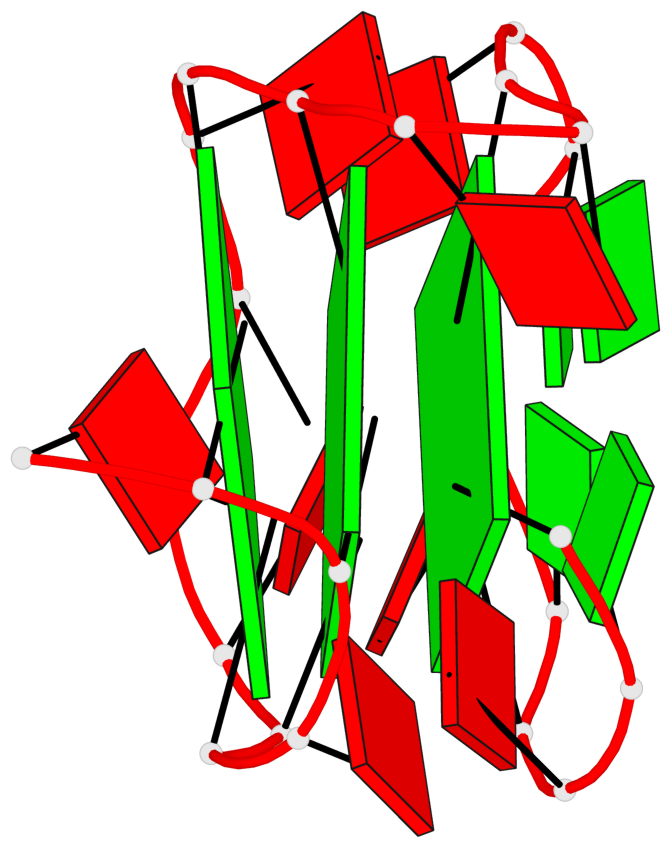
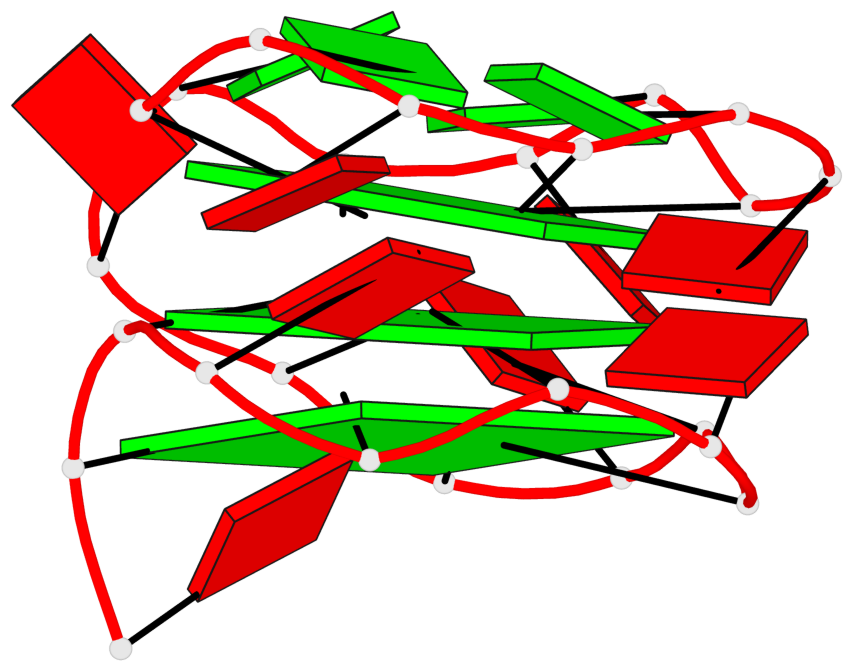
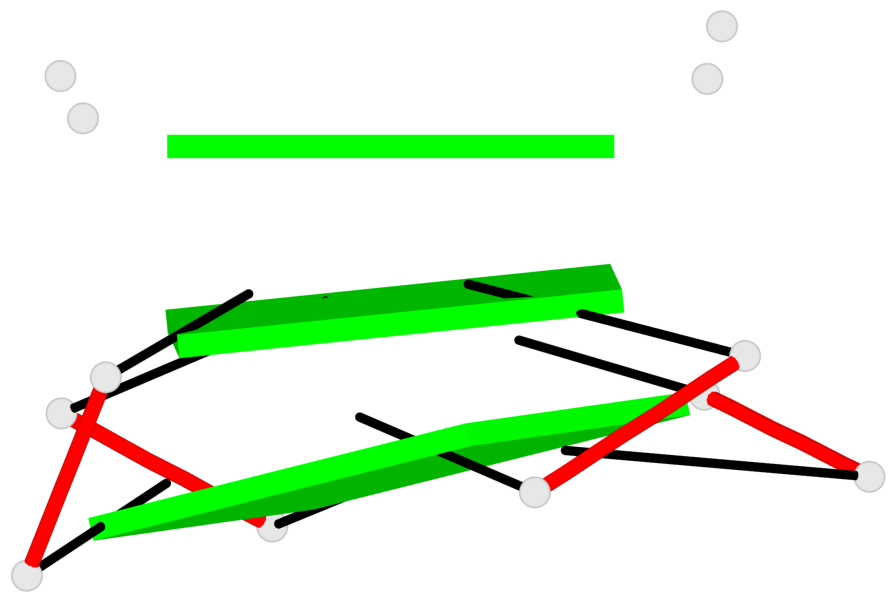
Base-block schematics in six views
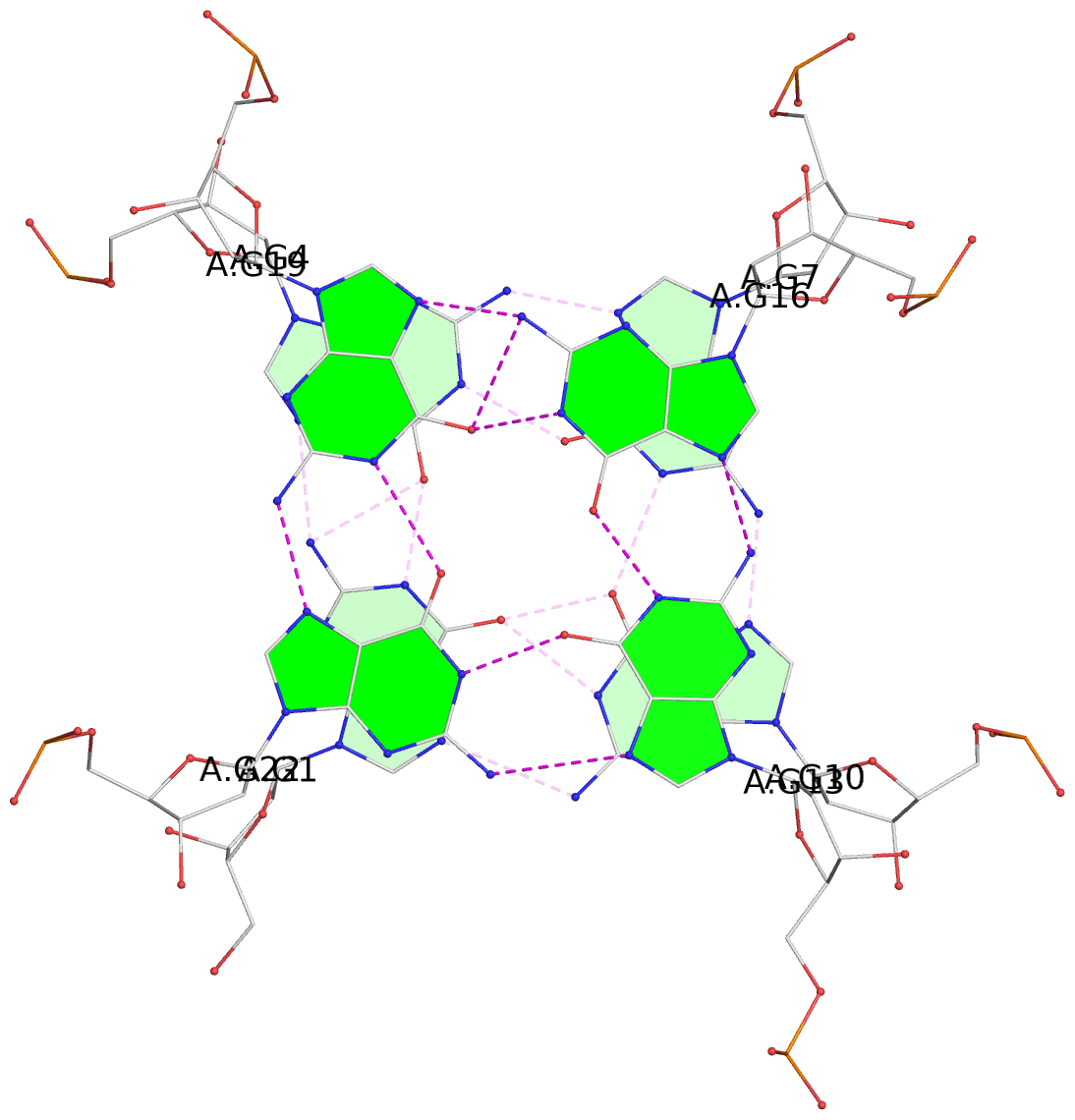
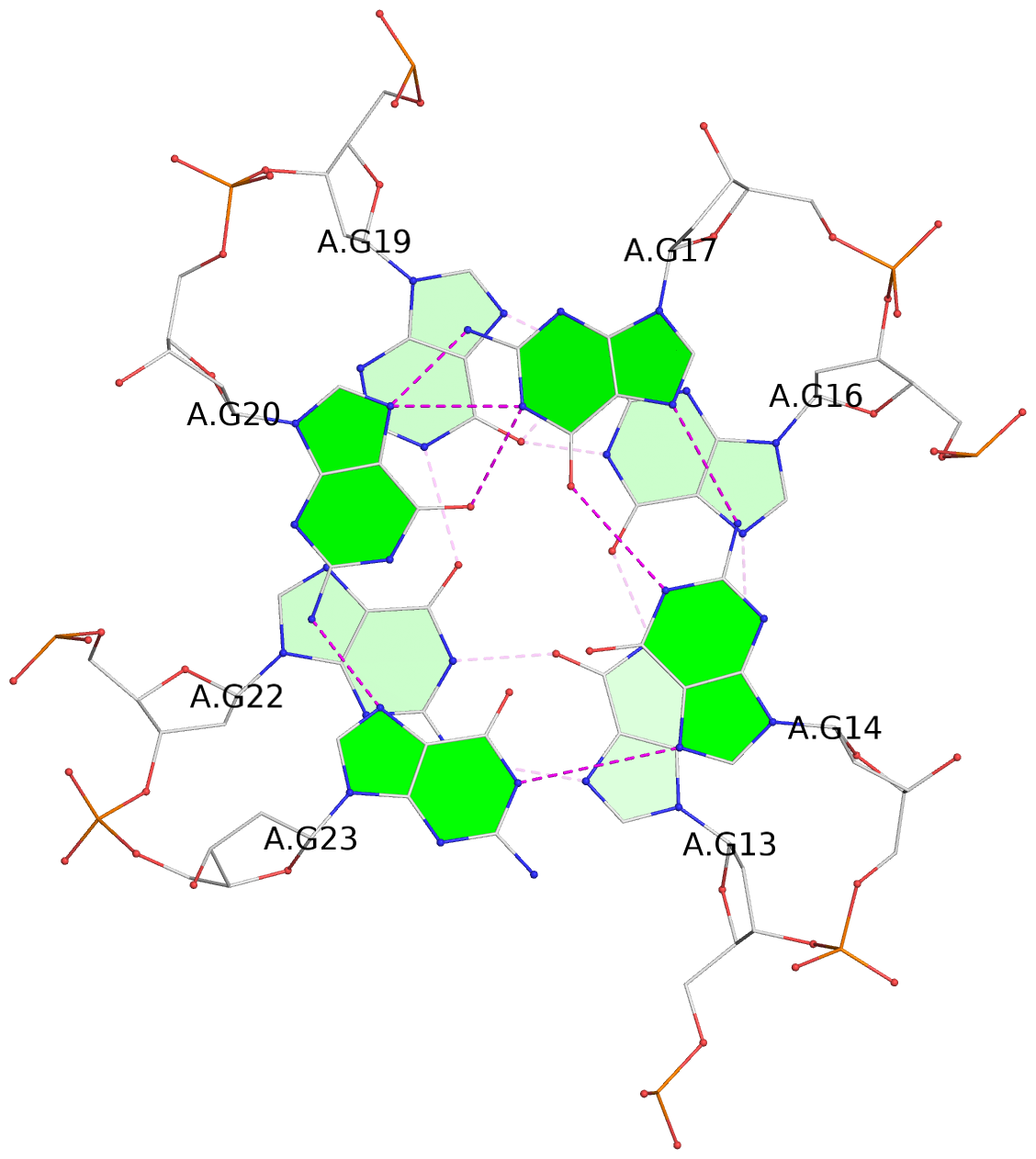
List of 3 G-tetrads
1 glyco-bond=---- sugar=-... groove=---- planarity=0.322 type=other nts=4 GGGG A.DG1,A.DG4,A.DG7,A.DG10 2 glyco-bond=---- sugar=.... groove=---- planarity=0.381 type=other nts=4 GGGG A.DG13,A.DG16,A.DG19,A.DG22 3 glyco-bond=---- sugar=---. groove=---- planarity=0.522 type=bowl nts=4 GGGG A.DG14,A.DG17,A.DG20,A.DG23
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 2 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 2(-P-P-P), parallel(4+0)
List of 3 non-stem G4-loops (including the two closing Gs)
1 type=lateral helix=#1 nts=4 GGAG A.DG1,A.DG2,A.DA3,A.DG4 2 type=lateral helix=#1 nts=4 GGAG A.DG4,A.DG5,A.DA6,A.DG7 3 type=lateral helix=#1 nts=4 GGAG A.DG7,A.DG8,A.DA9,A.DG10