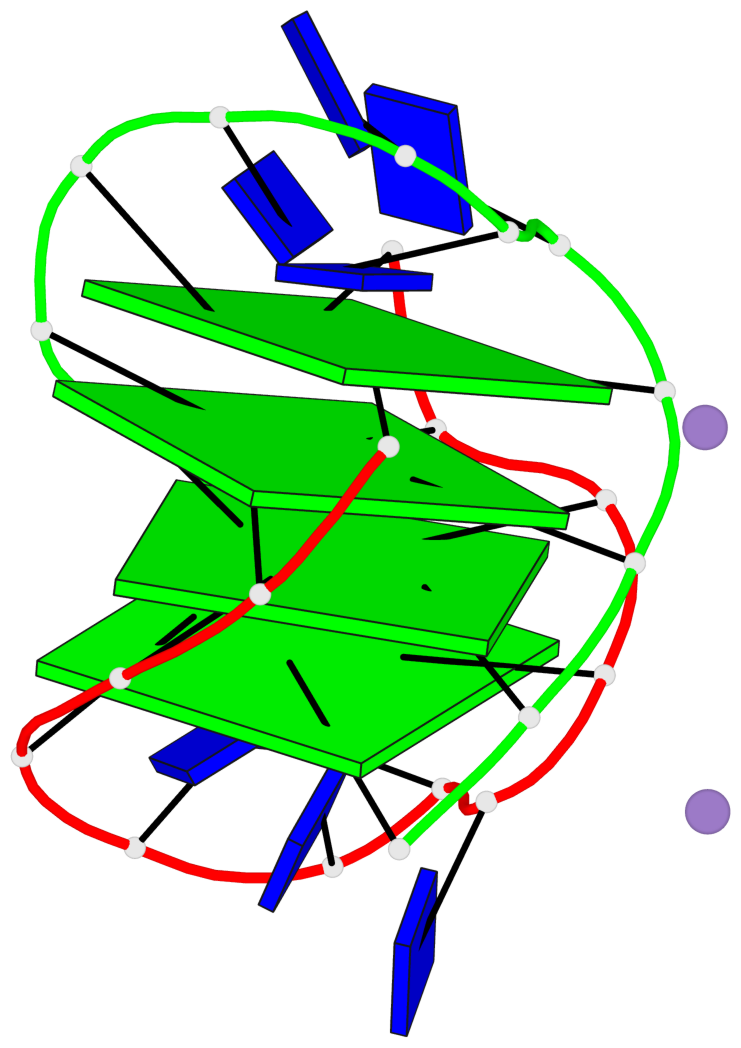
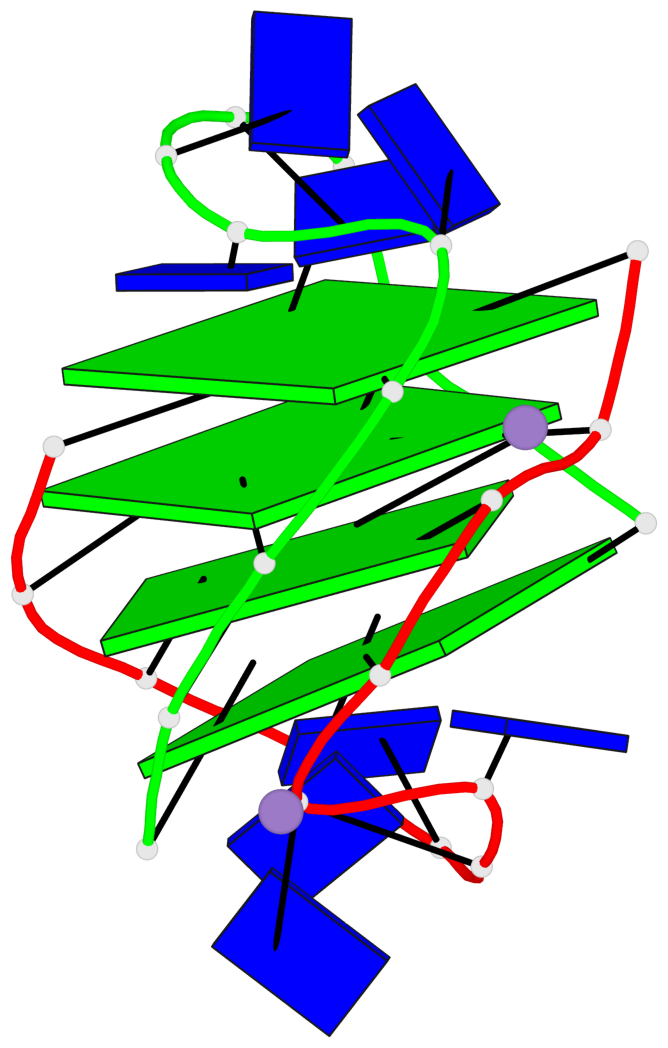
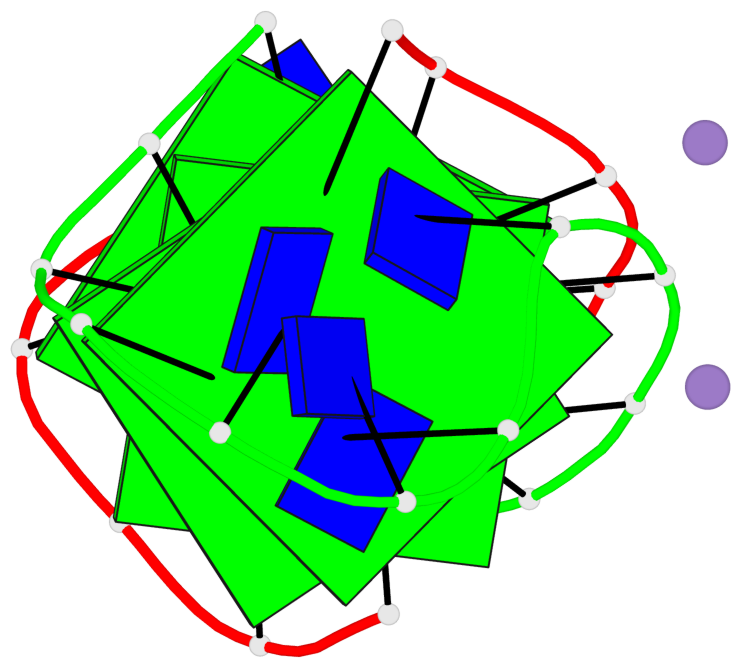
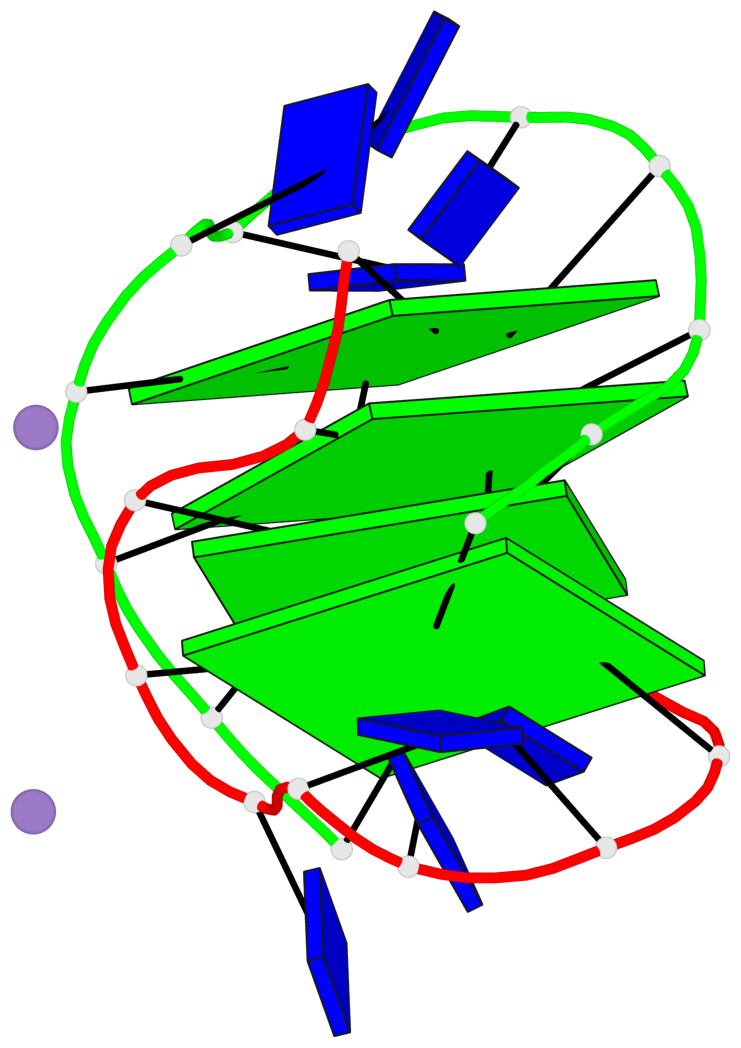
Detailed DSSR results for the G-quadruplex: PDB entry 1qdk
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 1qdk
- Class
- DNA
- Method
- NMR
- Summary
- The NMR study of DNA quadruplex structure, (12mer) DNA
- Reference
- Marathias VM, Wang KY, Kumar S, Pham TQ, Swaminathan S, Bolton PH (1996): "Determination of the number and location of the manganese binding sites of DNA quadruplexes in solution by EPR and NMR in the presence and absence of thrombin." J.Mol.Biol., 260, 378-394. doi: 10.1006/jmbi.1996.0408.
- Abstract
- The interaction of a DNA quadruplex with thrombin has been studied by first determining the sites of manganese binding to the quadruplex in the absence of thrombin. This has been followed by determining if the interactions with thrombin displace the bound manganese. A different DNA quadruplex has also been studied as a control. The refined solution structures of two DNA quadruplexes have been used to predict the electrostatic potentials of these DNAs. The calculated electrostatic potentials have been used to predict the locations of the binding sites of the paramagnetic ion manganese to these DNAs. The enhanced relaxation of DNA protons due to the binding of the paramagnetic metal ion Mn2+ has been used to experimentally determine the locations of the binding sites. The NMR results and the predictions based on the electrostatic potentials both place the binding sites of the manganese in the narrow grooves of these quadruplex DNAs. The predicted locations are spatially close to those experimentally observed, and the predicted and experimental locations also have similar electrostatic potential energy. These results have allowed a validation of the predictions of electrostatic potentials from structure. The 15mer quadruplex has two strong Mn2+ binding sites with one in each narrow groove. Both Mn2+ are released when the 15mer is complexed with thrombin, indicating that both narrow grooves are involved in the 15mer-thrombin interactions. The dimer quadruplex has a different structural motif than the 15mer and the presence of thrombin does not appreciably affect its interactions with Mn2+.
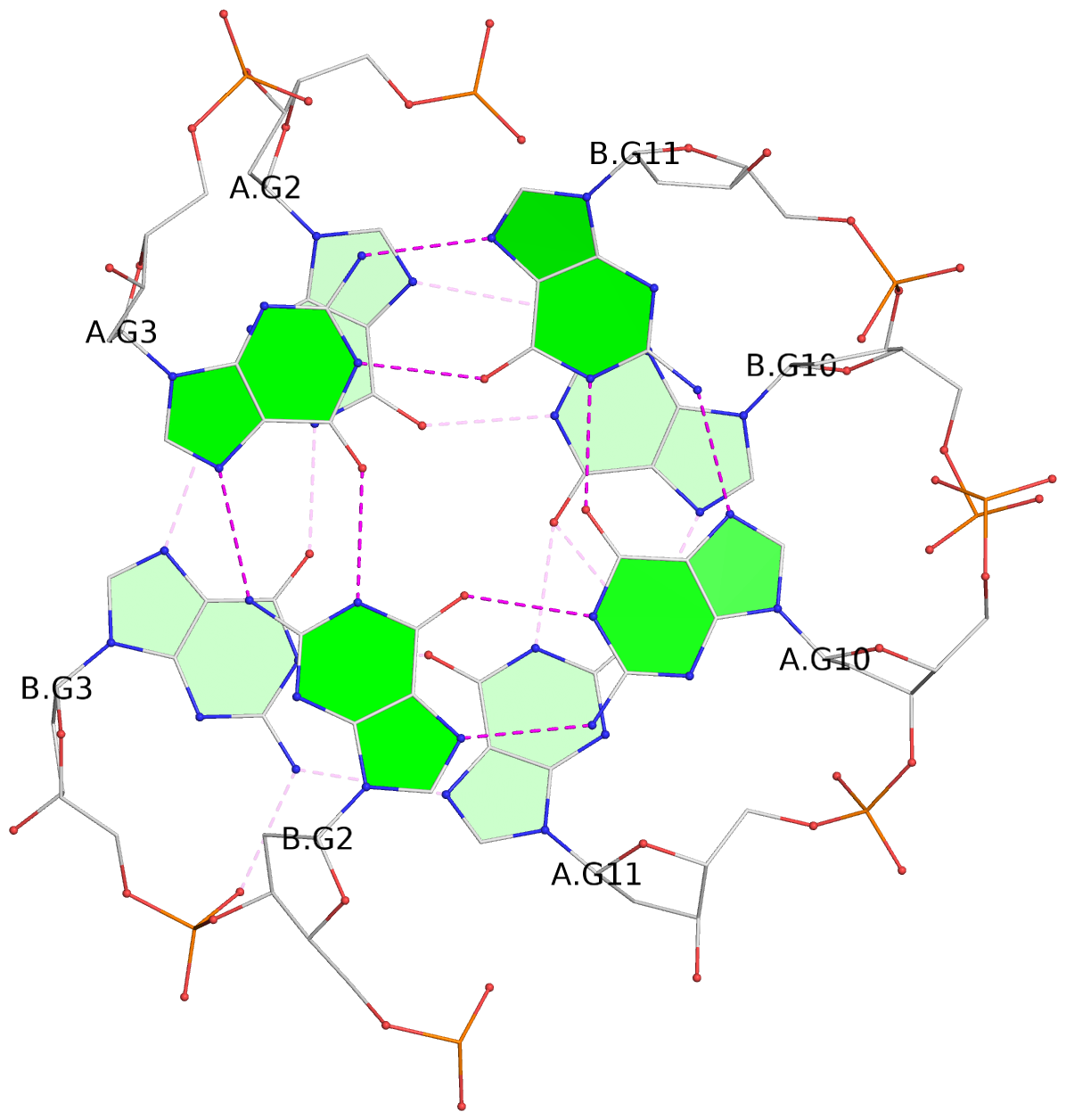
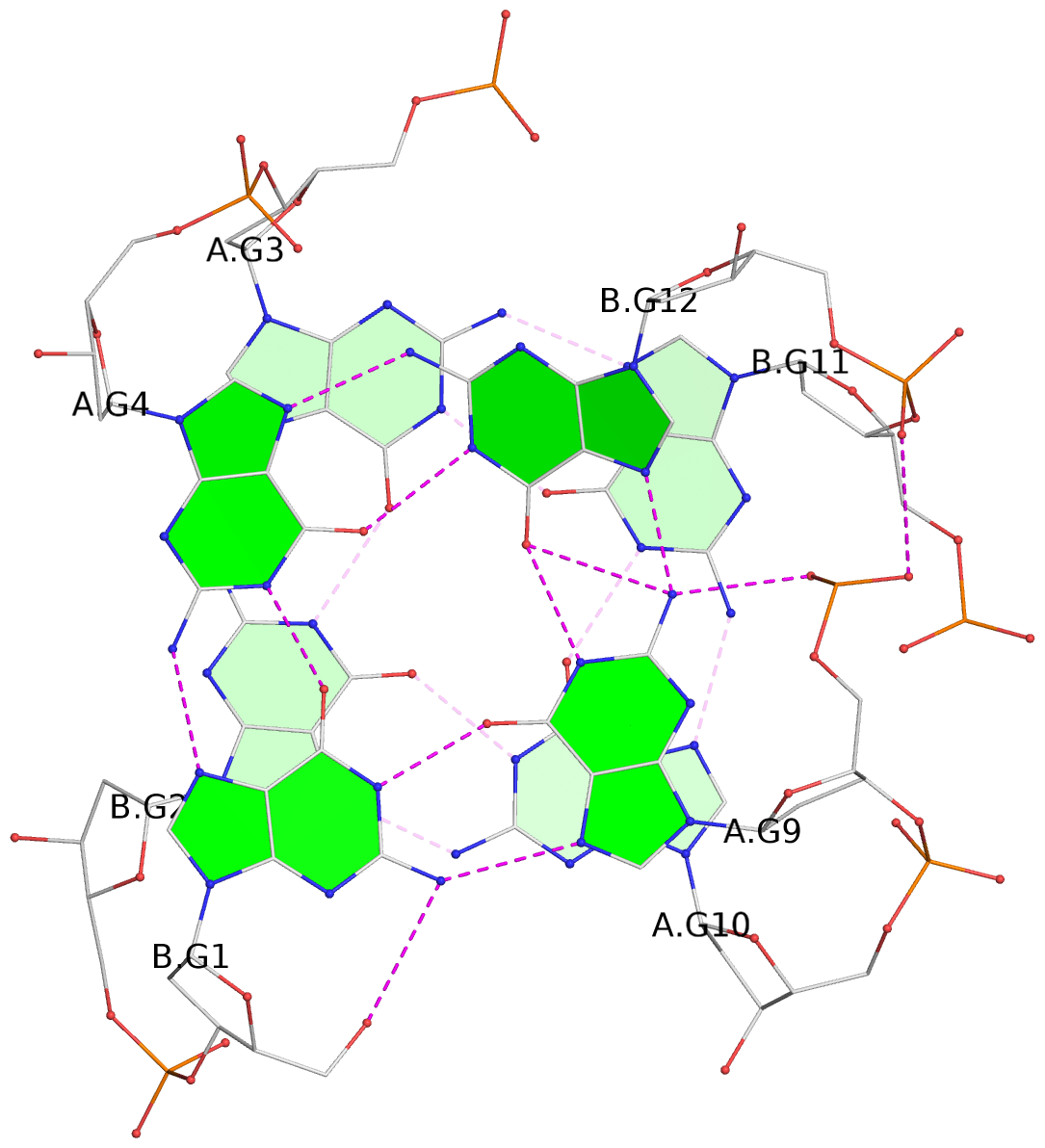
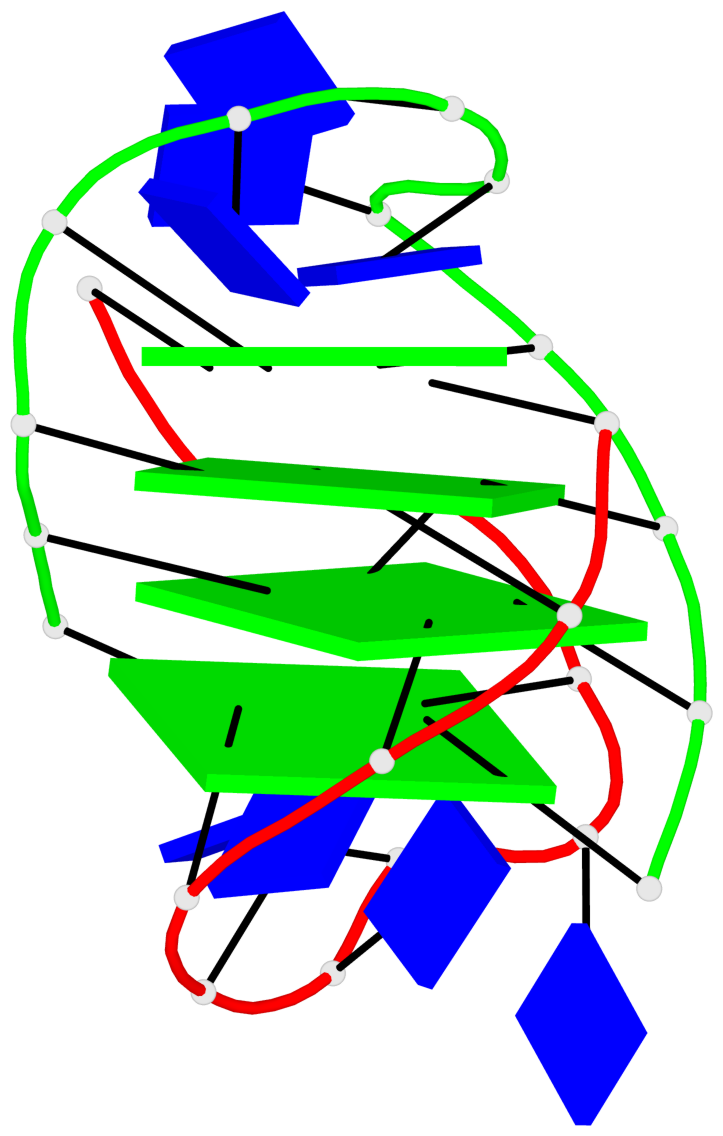
- G4 notes
- 4 G-tetrads, 1 G4 helix, 1 G4 stem, (2+2), UDDU
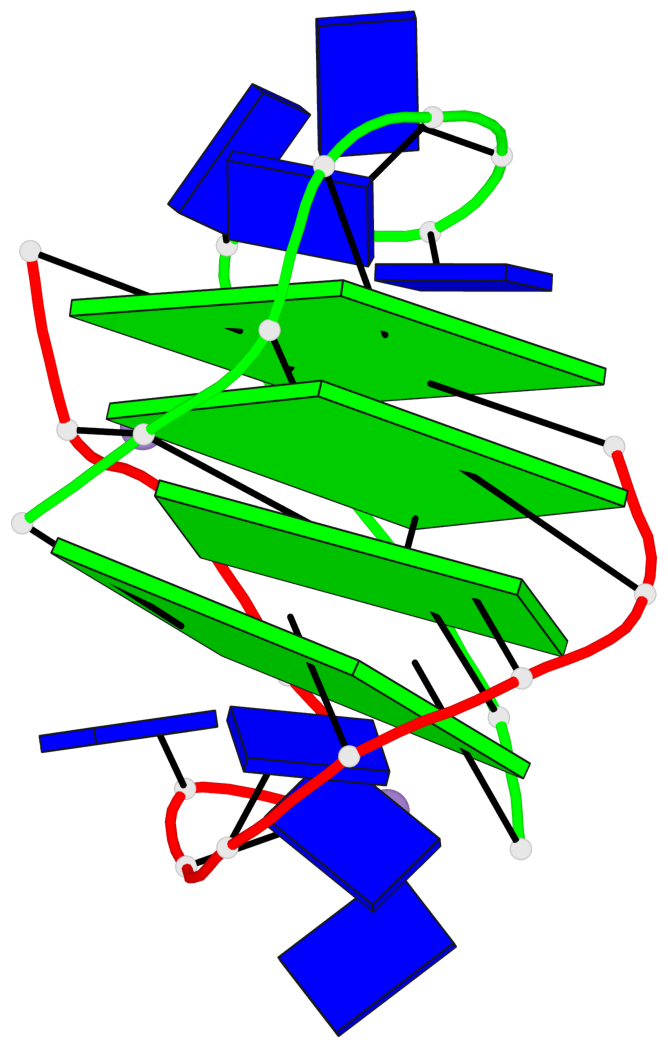
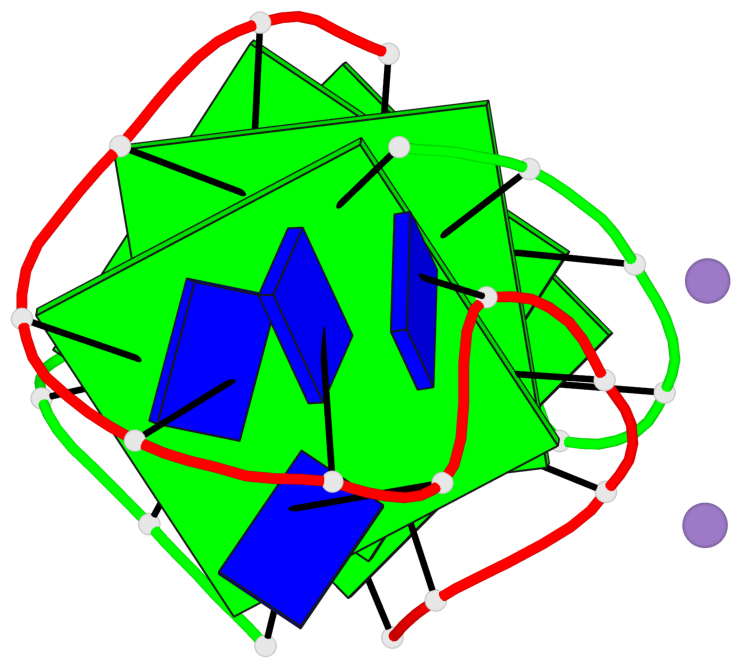
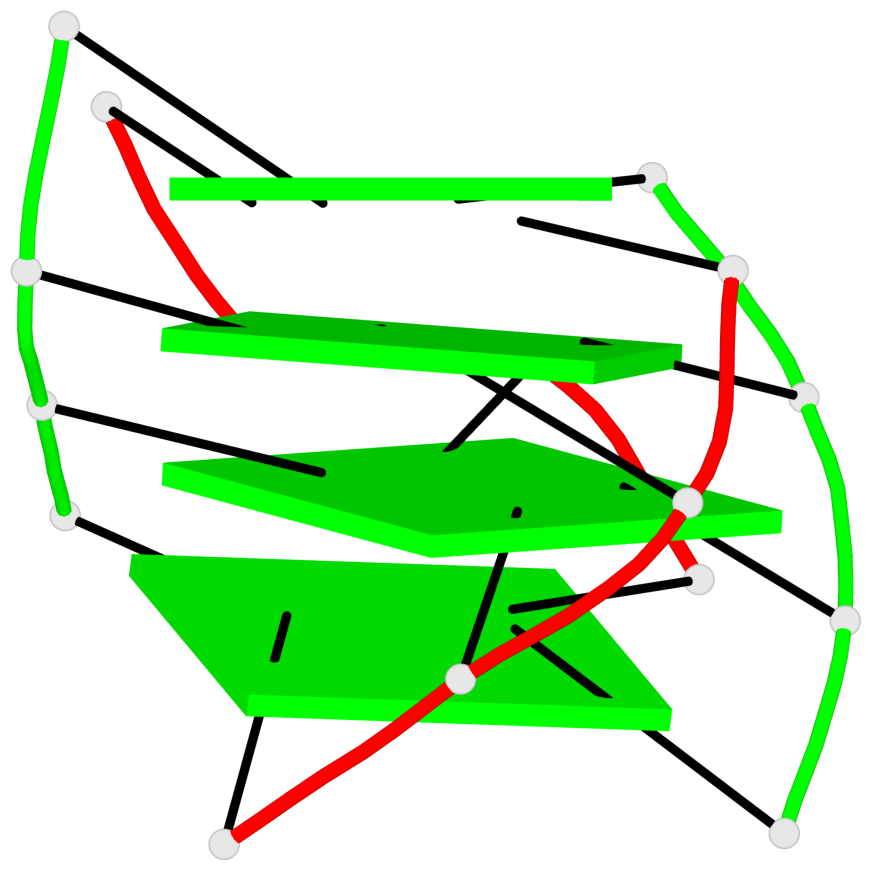
Base-block schematics in six views
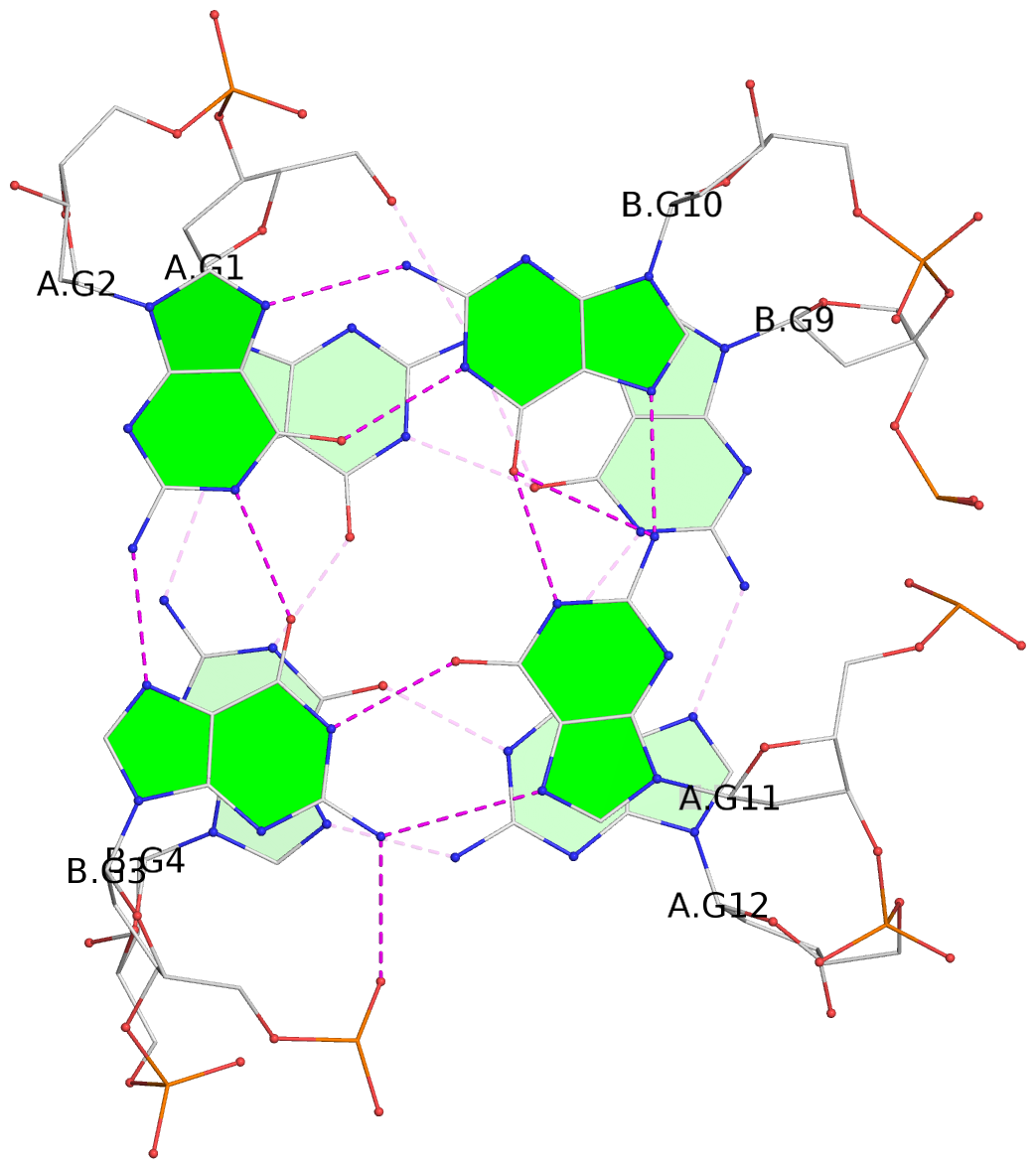
List of 4 G-tetrads
1 glyco-bond=s--s sugar=33.- groove=w-n- planarity=0.480 type=bowl nts=4 GGGG A.DG1,B.DG4,A.DG12,B.DG9 2 glyco-bond=-ss- sugar=3.-. groove=w-n- planarity=0.414 type=other nts=4 GGGG A.DG2,B.DG3,A.DG11,B.DG10 3 glyco-bond=s--s sugar=33-. groove=w-n- planarity=0.397 type=other nts=4 GGGG A.DG3,B.DG2,A.DG10,B.DG11 4 glyco-bond=-ss- sugar=.3-. groove=w-n- planarity=0.562 type=bowl nts=4 GGGG A.DG4,B.DG1,A.DG9,B.DG12
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 4 G-tetrad layers, inter-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.