Detailed DSSR results for the G-quadruplex: PDB entry 1s47
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 1s47
- Class
- DNA
- Method
- X-ray (2.5 Å)
- Summary
- Crystal structure analysis of the DNA quadruplex d(tggggt)s2
- Reference
- Caceres C, Wright G, Gouyette C, Parkinson G, Subirana JA (2004): "A Thymine tetrad in d(TGGGGT) quadruplexes stabilized with Tl+1/Na+1 ions." Nucleic Acids Res., 32, 1097-1102. doi: 10.1093/nar/gkh269.
- Abstract
- We report two new structures of the quadruplex d(TGGGGT)4 obtained by single crystal X-ray diffraction. In one of them a thymine tetrad is found. Thus the yeast telomere sequences d(TG1-3) might be able to form continuous quadruplex structures, involving both guanine and thymine tetrads. Our study also shows substantial differences in the arrangement of thymines when compared with previous studies. We find five different types of organization: (i) groove binding with hydrogen bonds to guanines from a neighbour quadruplex; (ii) partially ordered groove binding, without any hydrogen bond; (iii) stacked thymine triads, formed at the 3'ends of the quadruplexes; (iv) a thymine tetrad between two guanine tetrads. Thymines are stabilized in pairs by single hydrogen bonds. A central sodium ion interacts with two thymines and contributes to the tetrad structure. (v) Completely disordered thymines which do not show any clear location in the crystal. The tetrads are stabilized by either Na+ or Tl+ ions. We show that by using MAD methods, Tl+ can be unambiguously located and distinguished from Na+. We can thus determine the preference for either ion in each ionic site of the structure under the conditions used by us.
- G4 notes
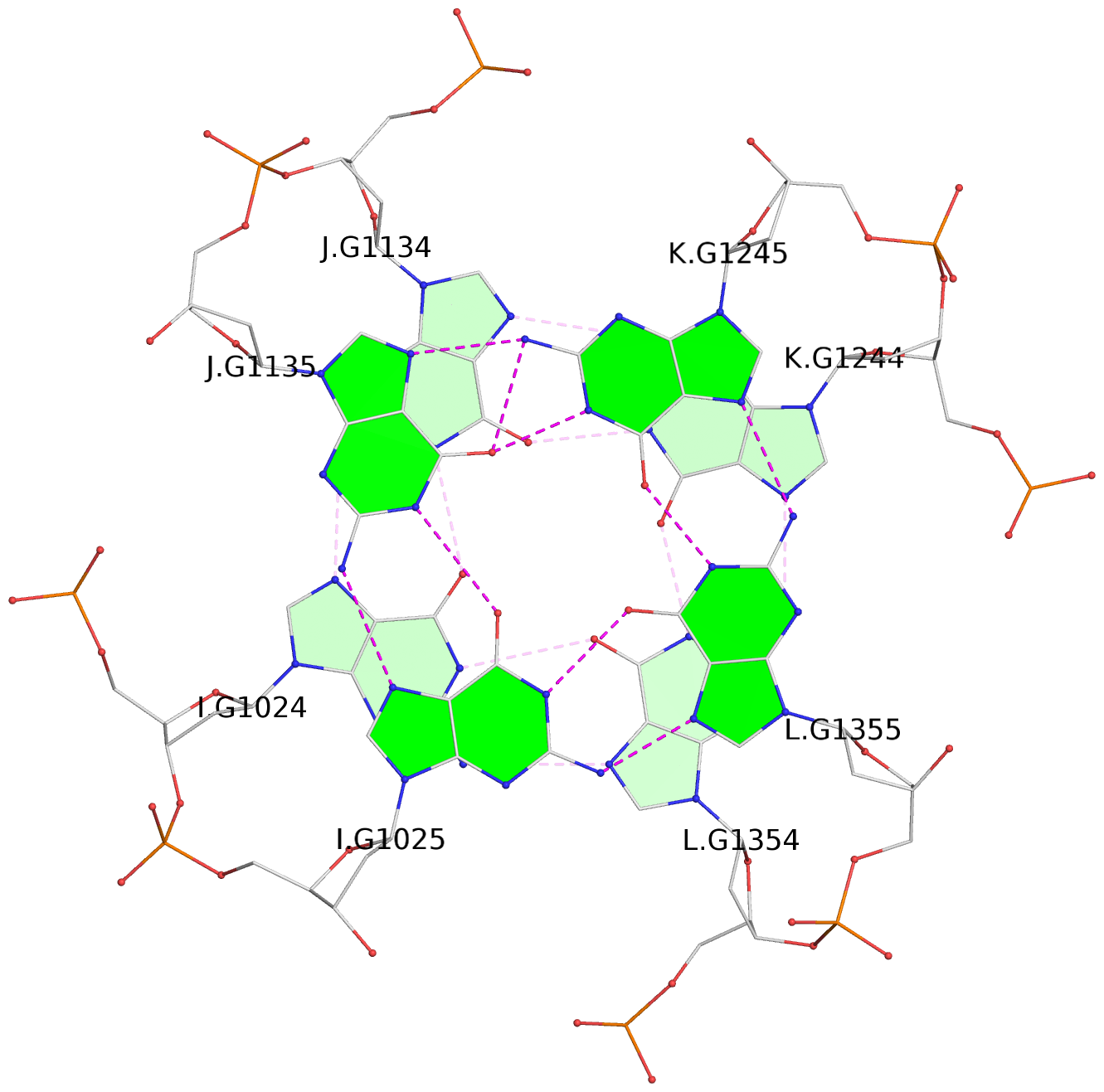
- 12 G-tetrads, 2 G4 helices, 3 G4 stems, 1 G4 coaxial stack, parallel(4+0), UUUU, coaxial interfaces: 5'/5'
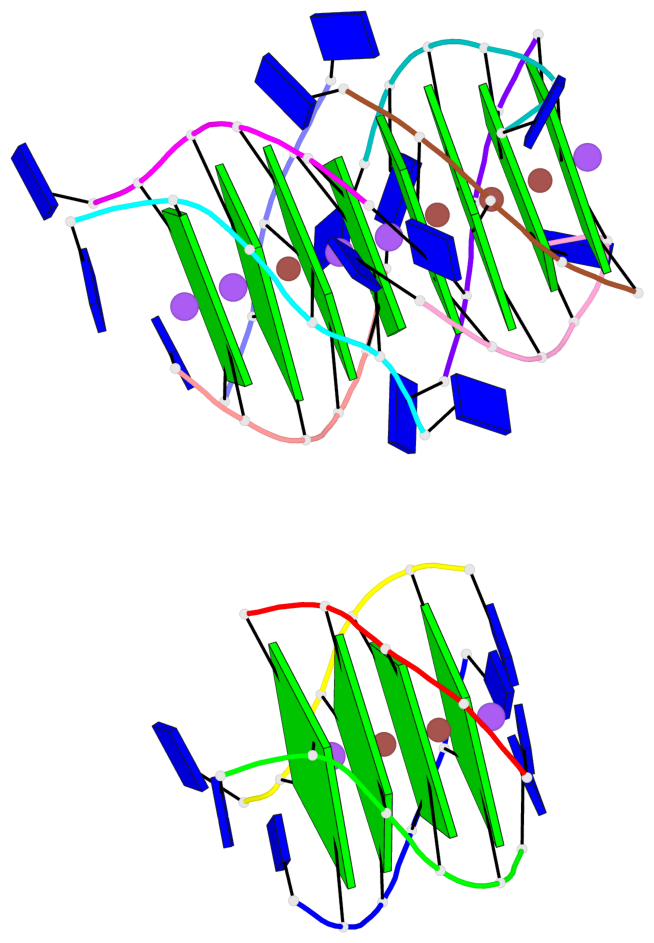
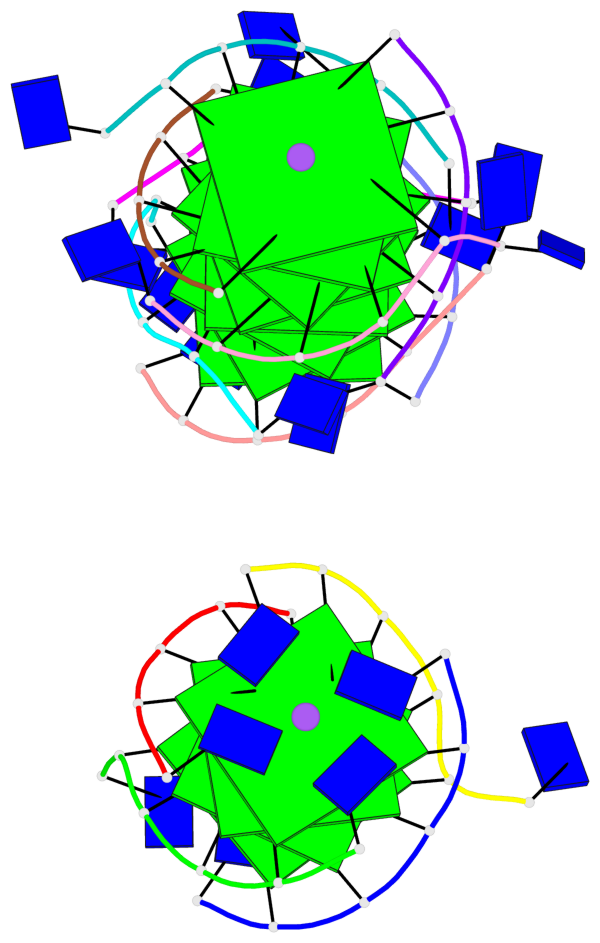
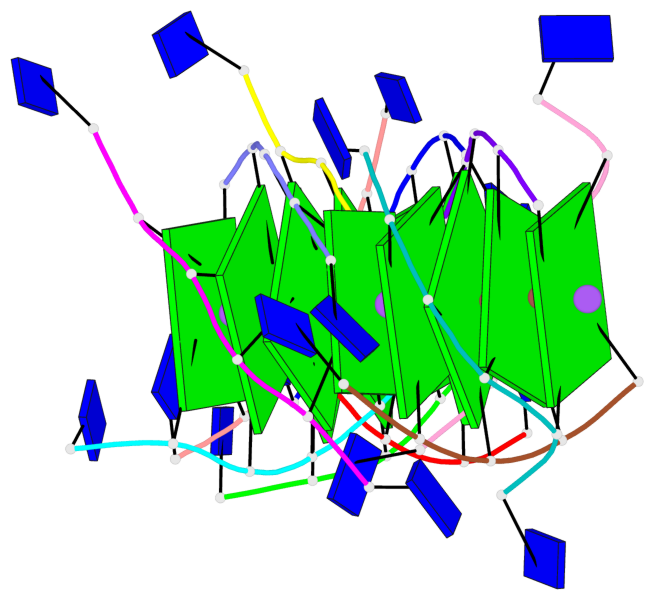
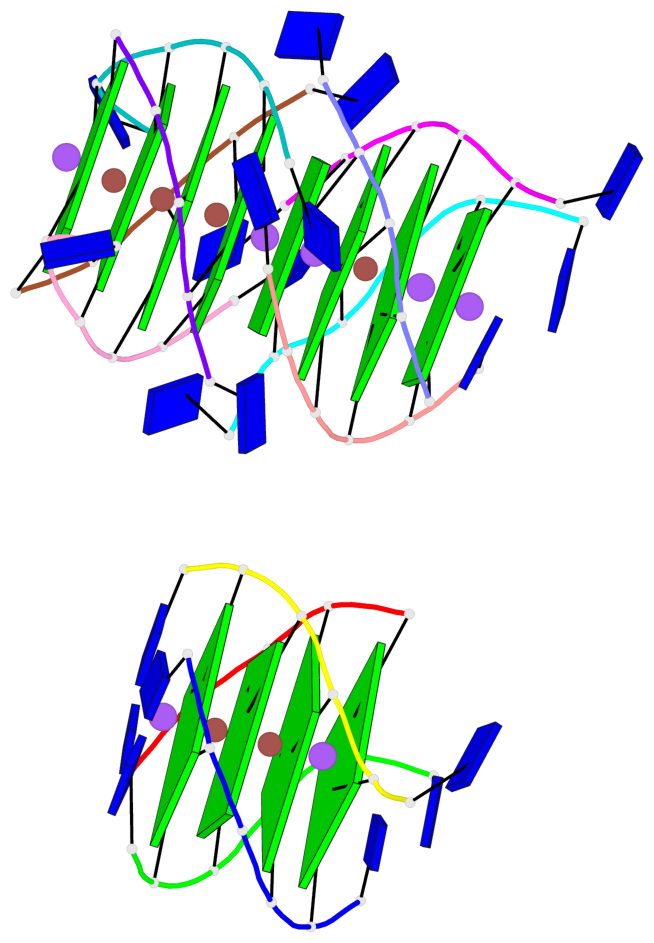
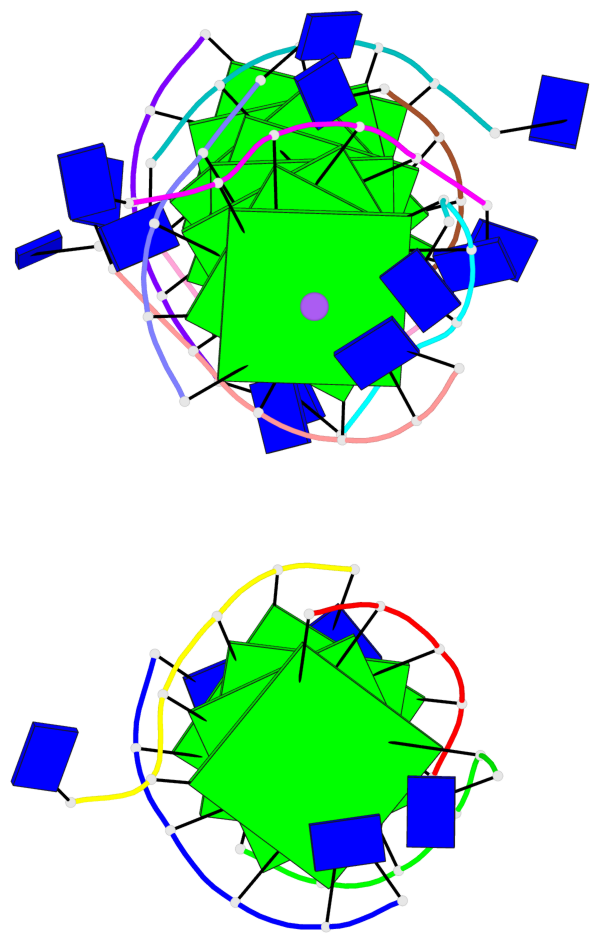
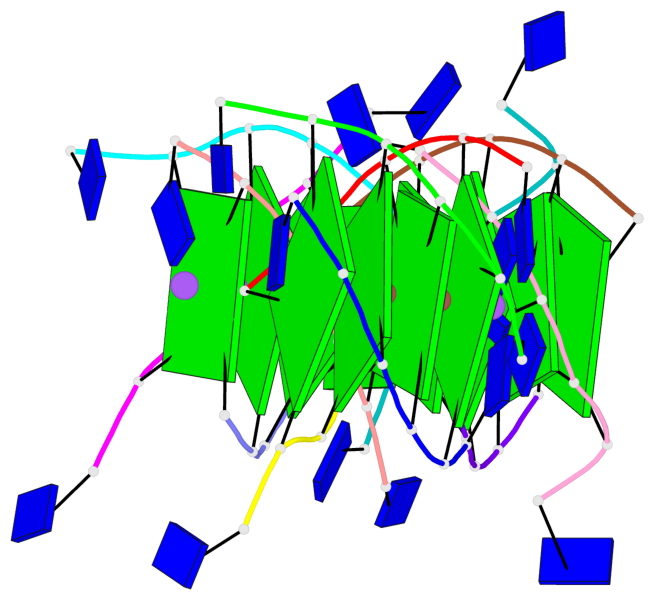
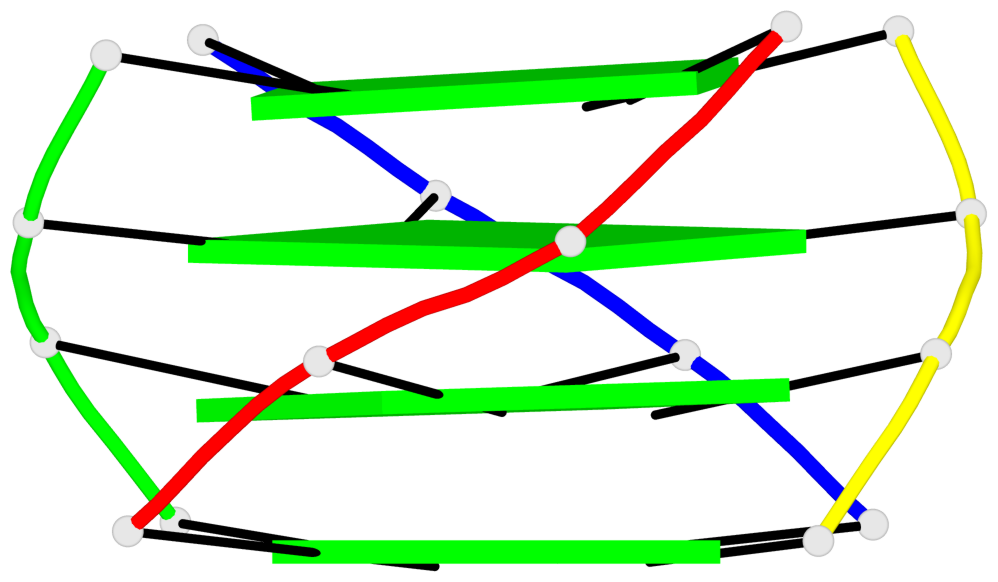
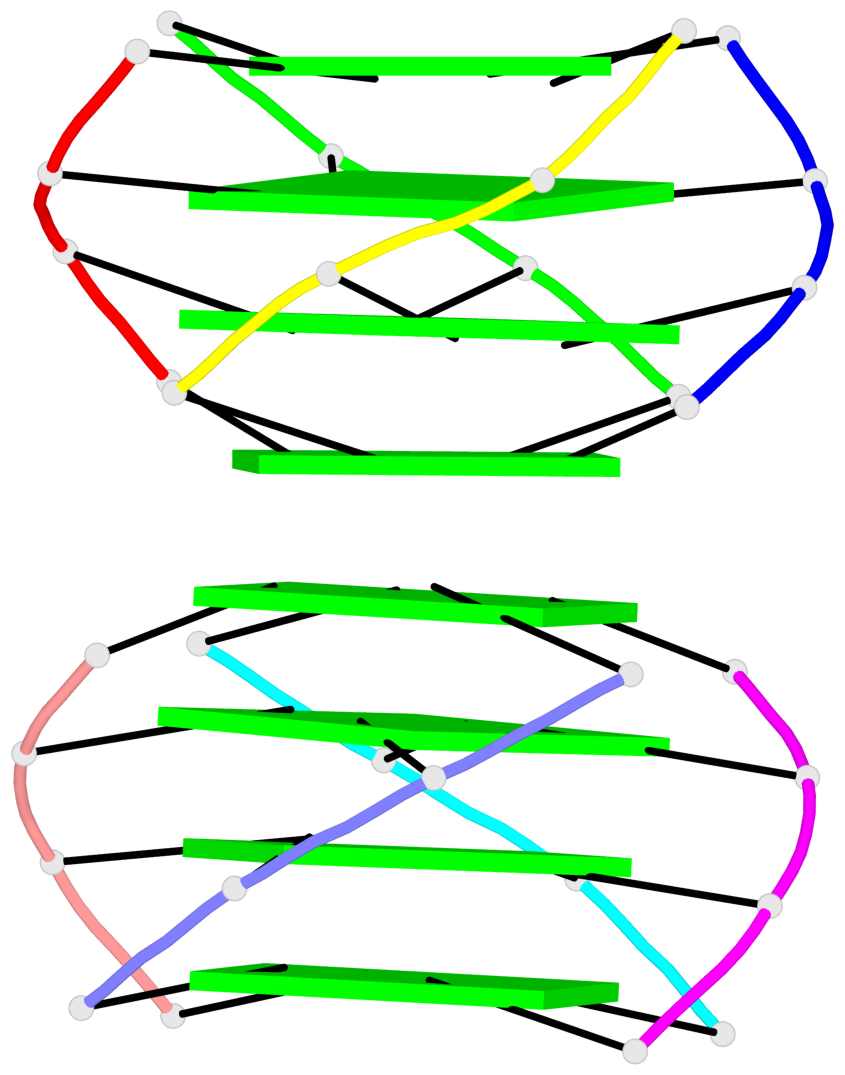
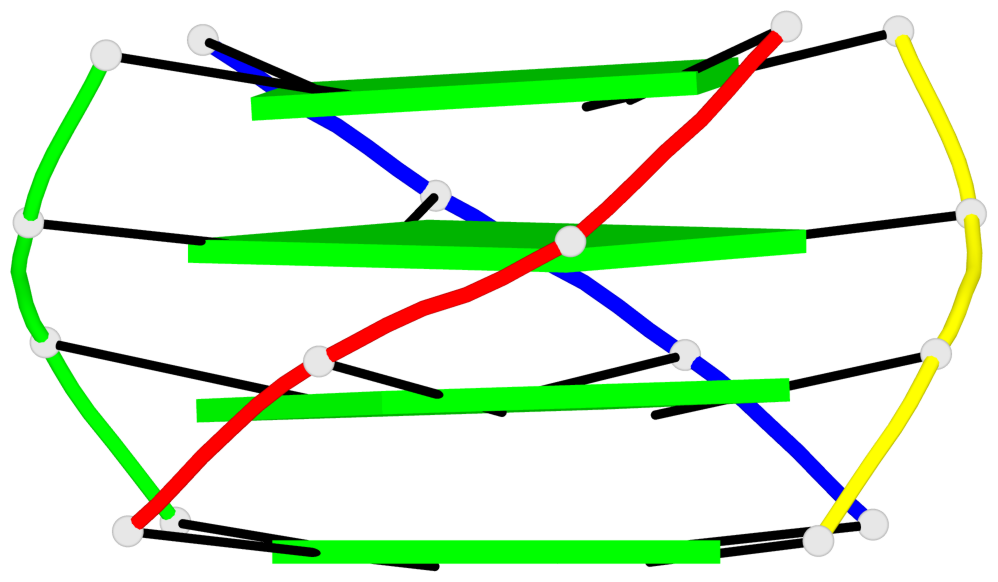
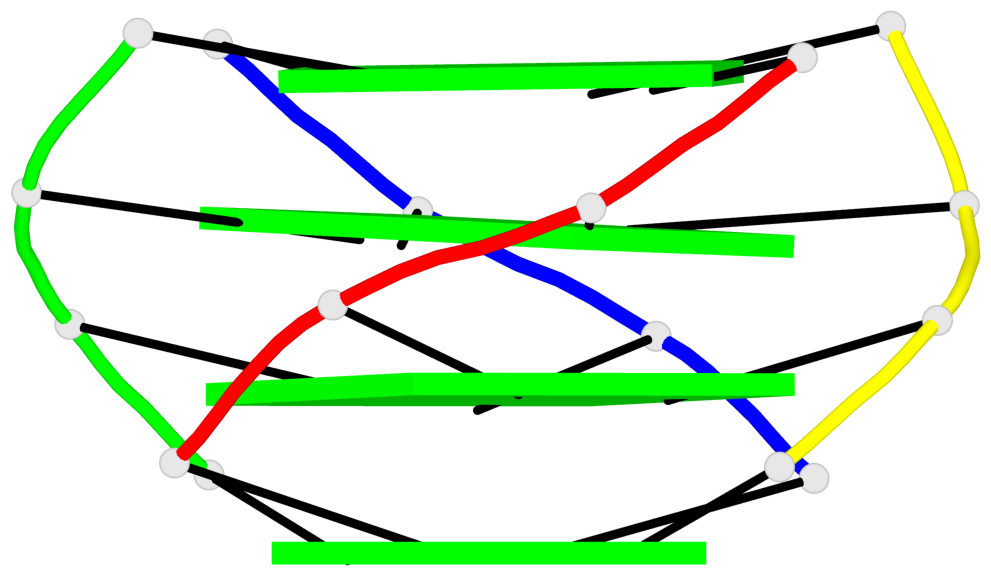
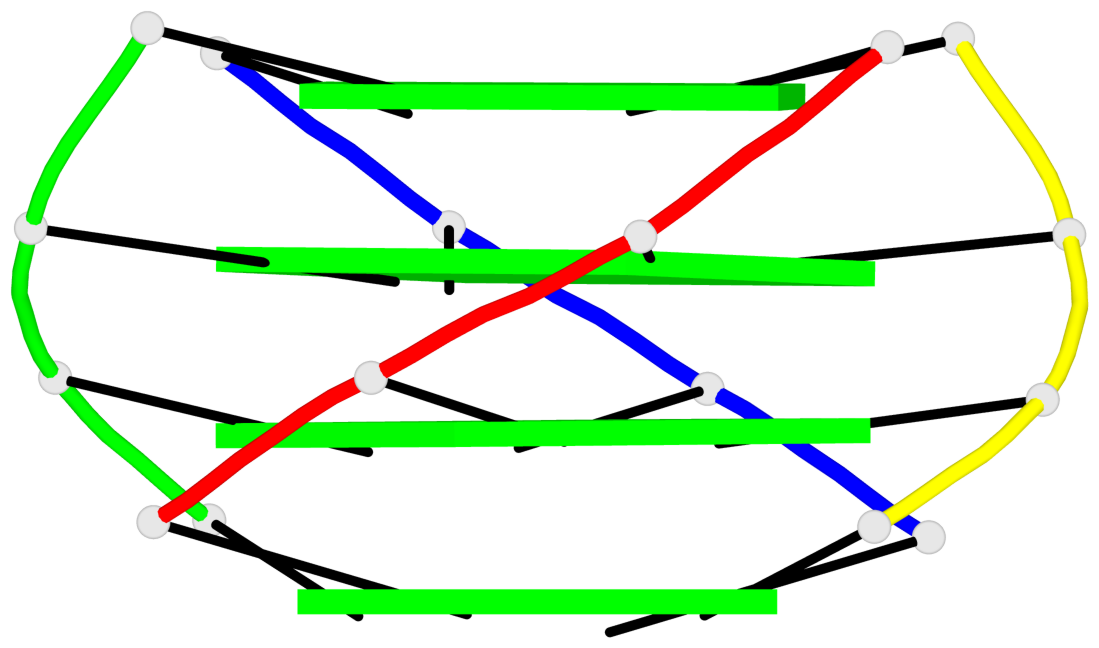
Base-block schematics in six views
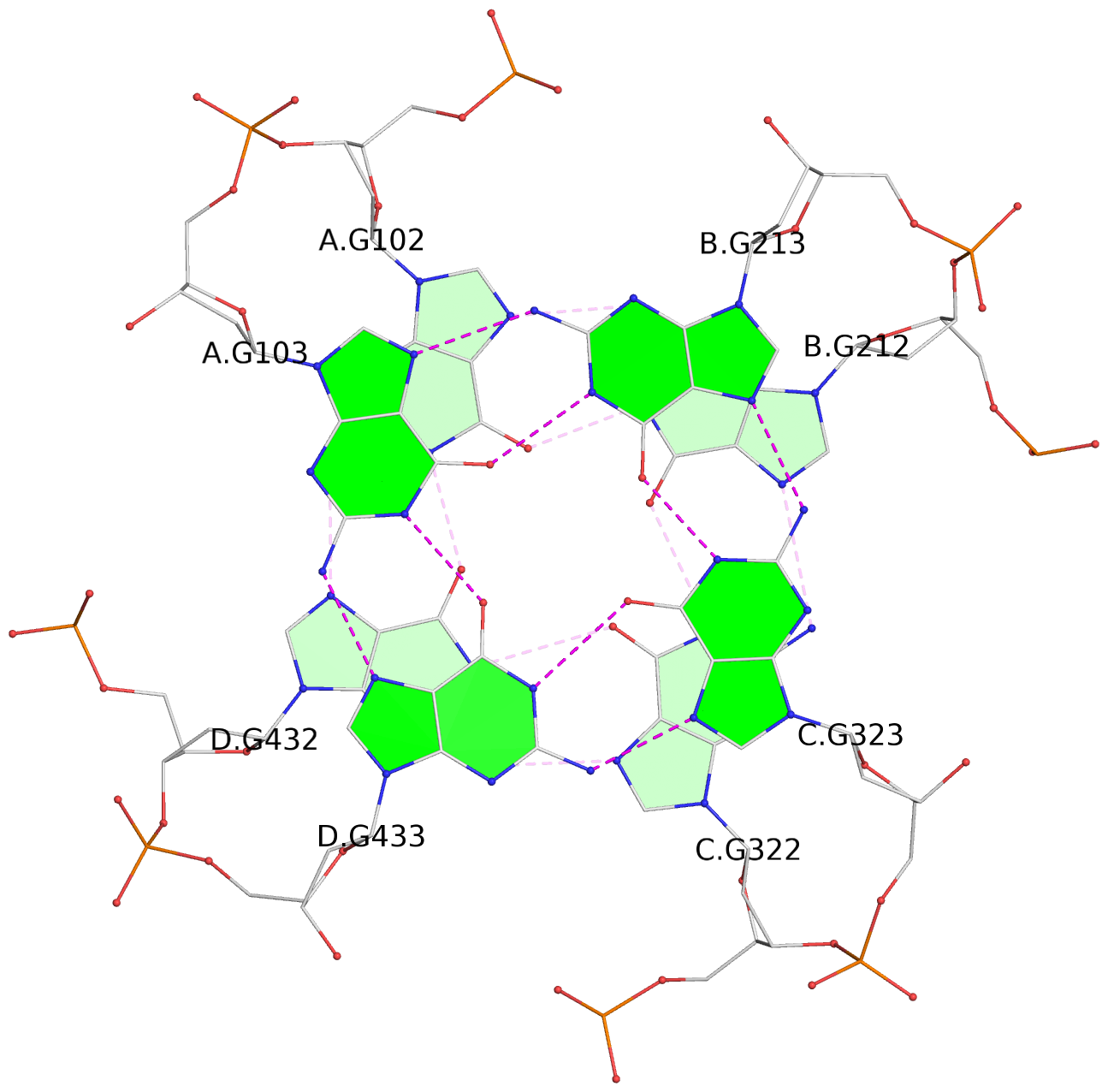
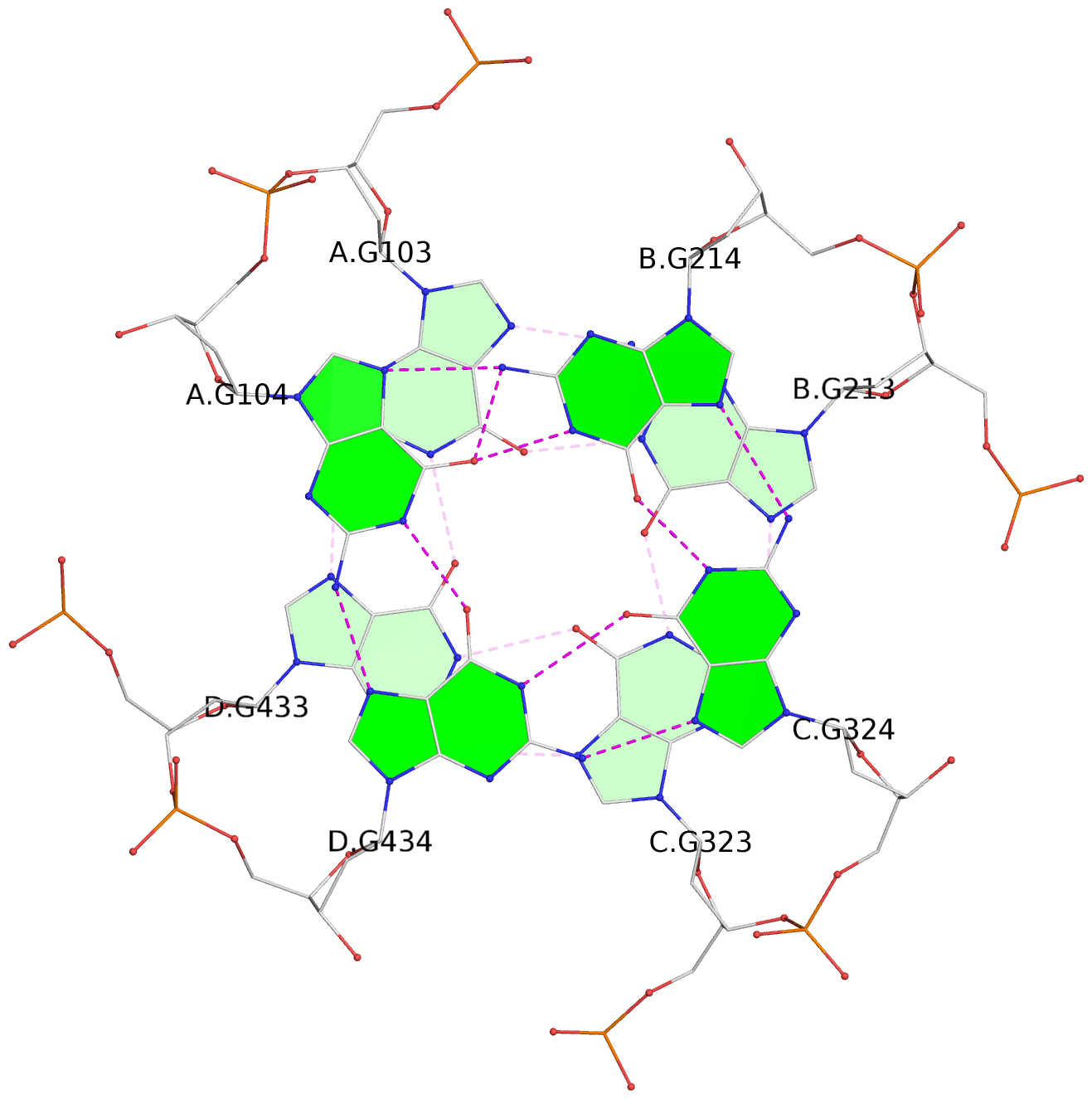
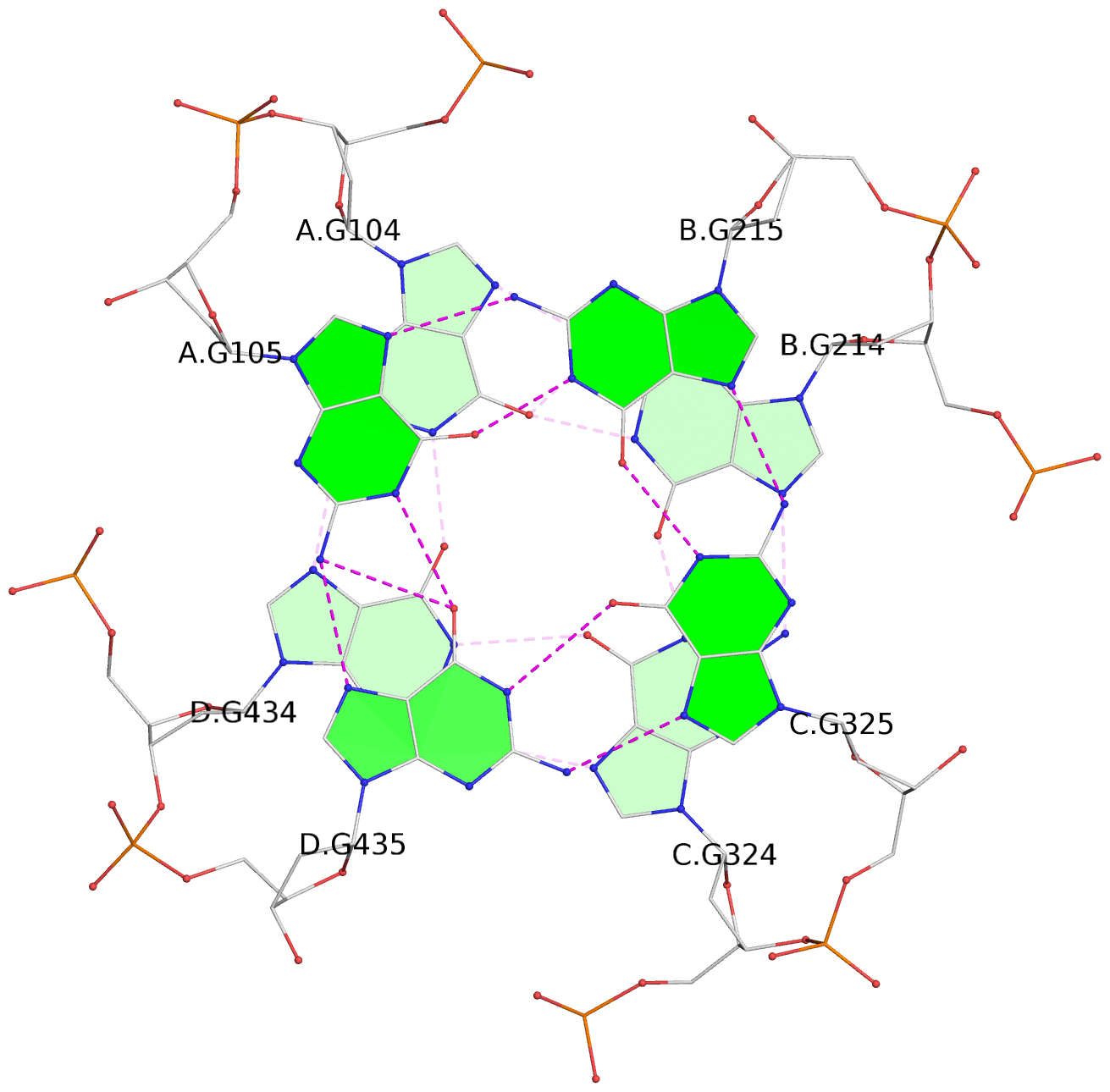
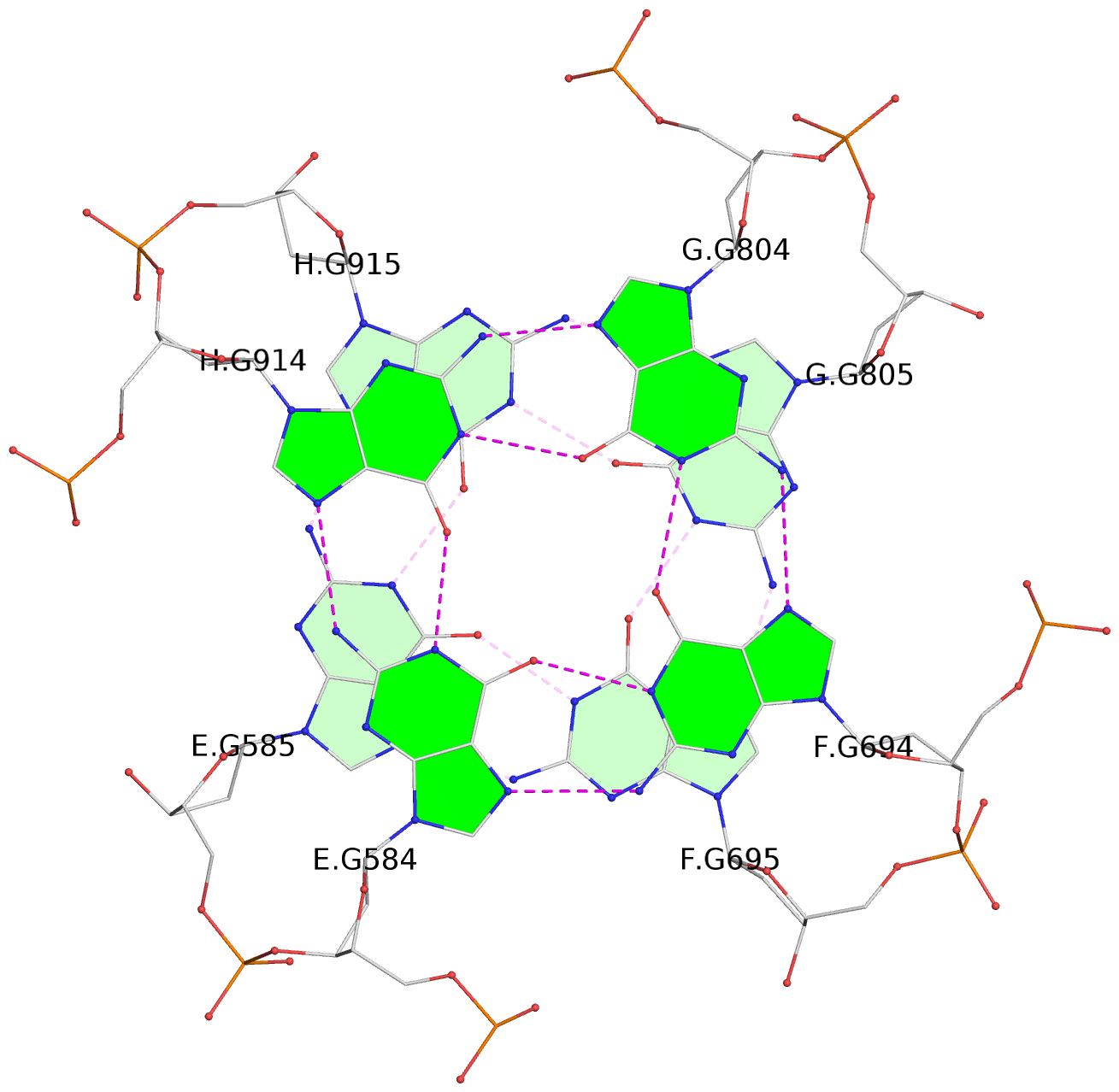
List of 12 G-tetrads
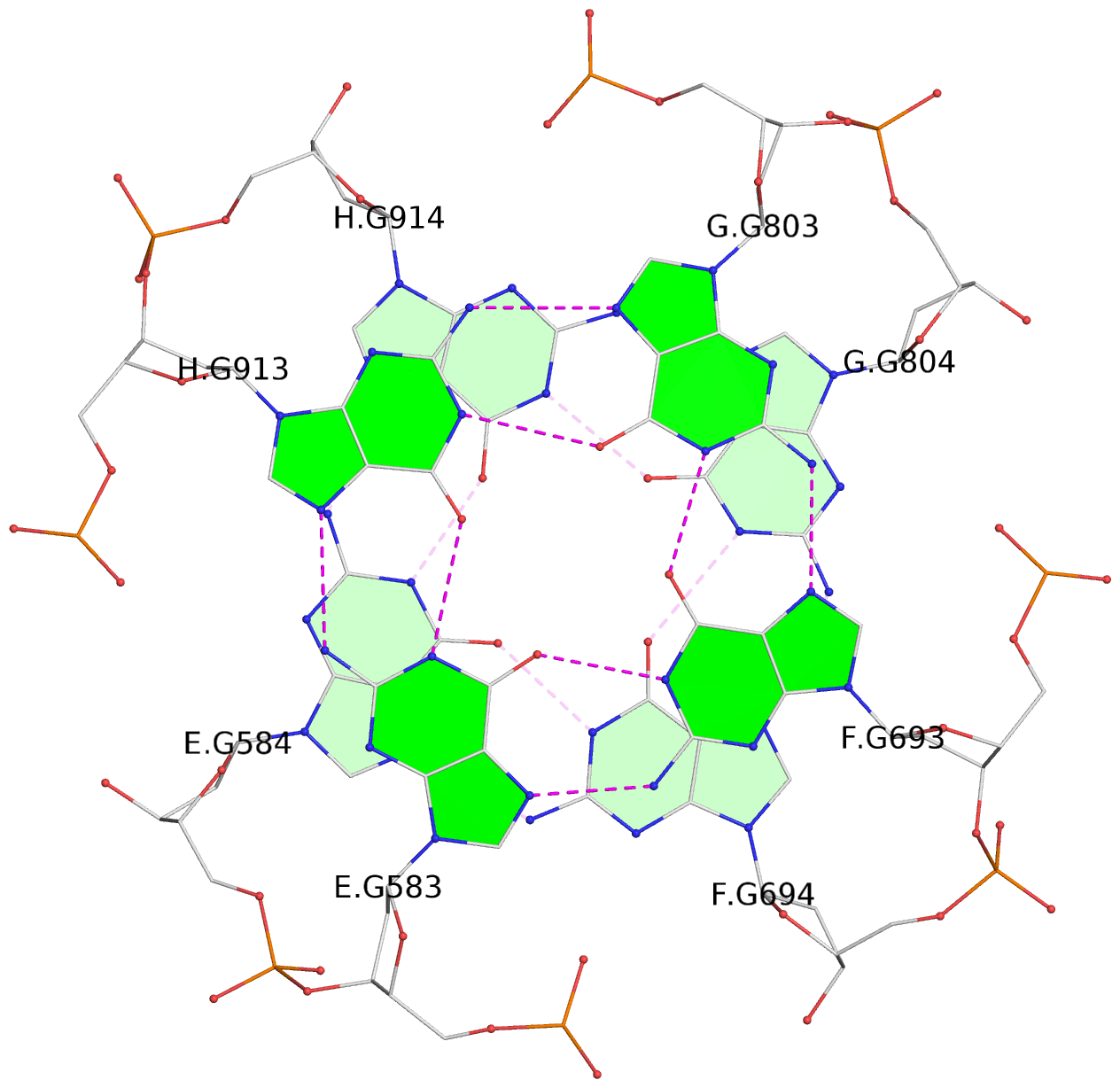
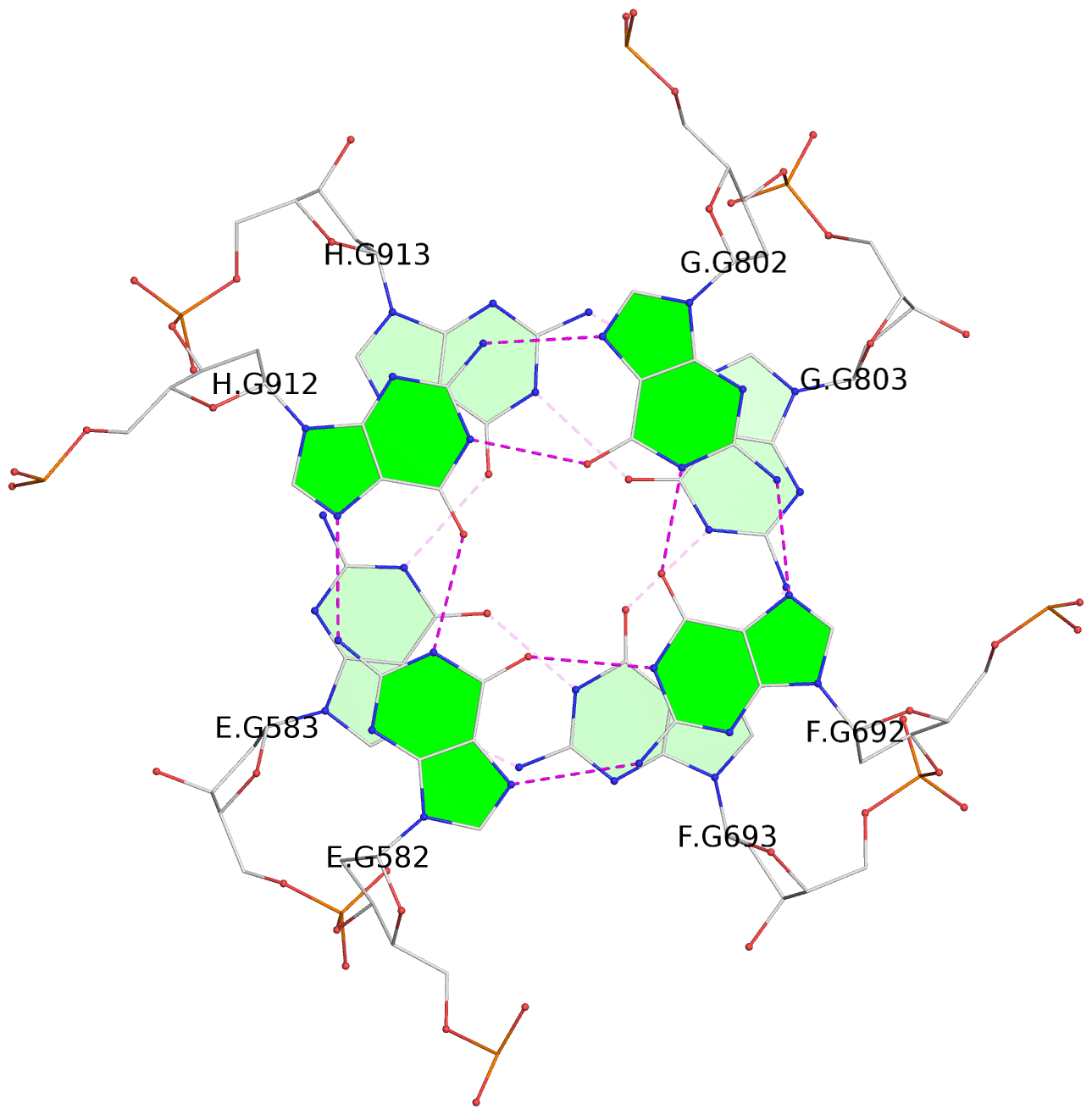
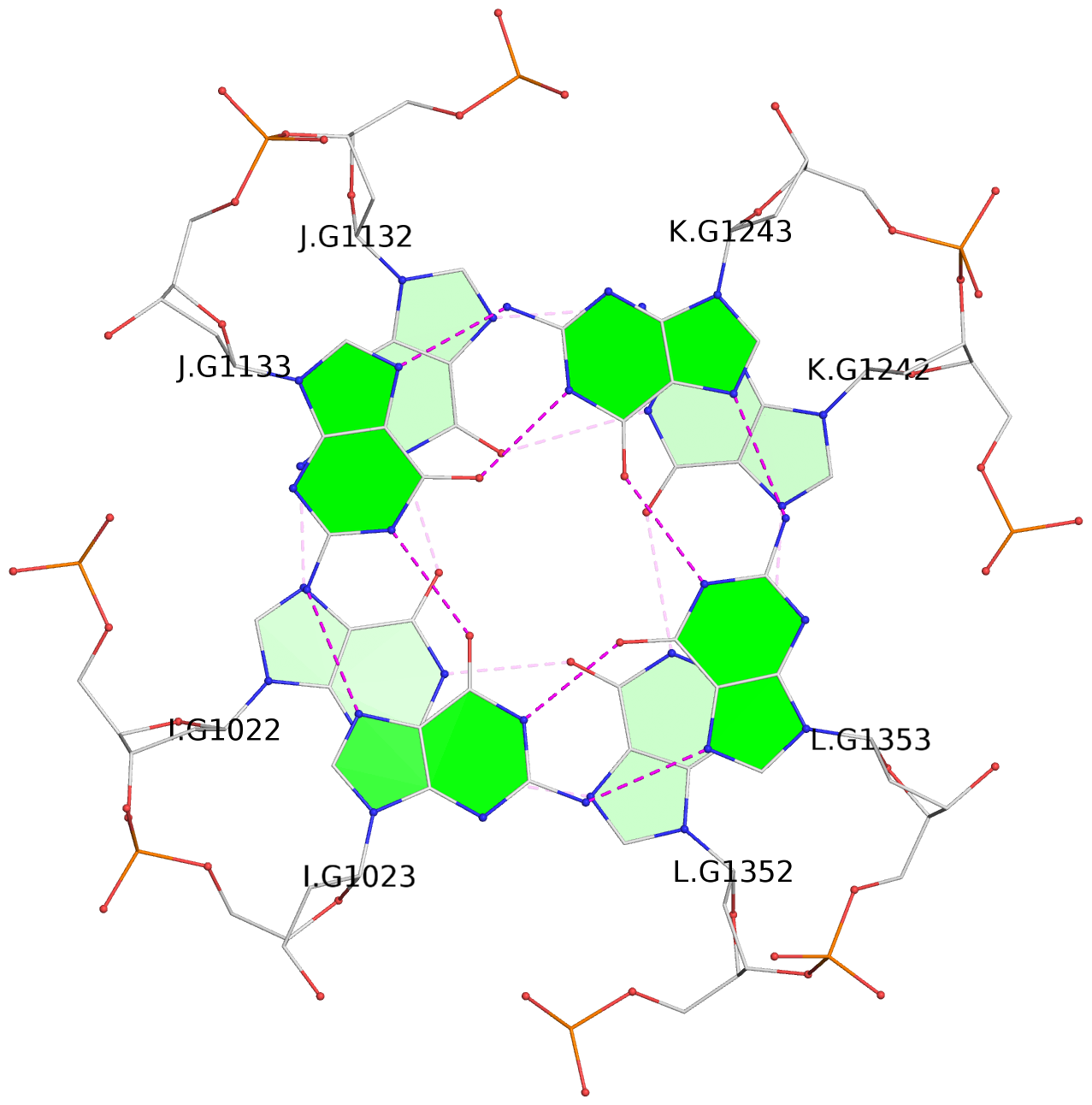
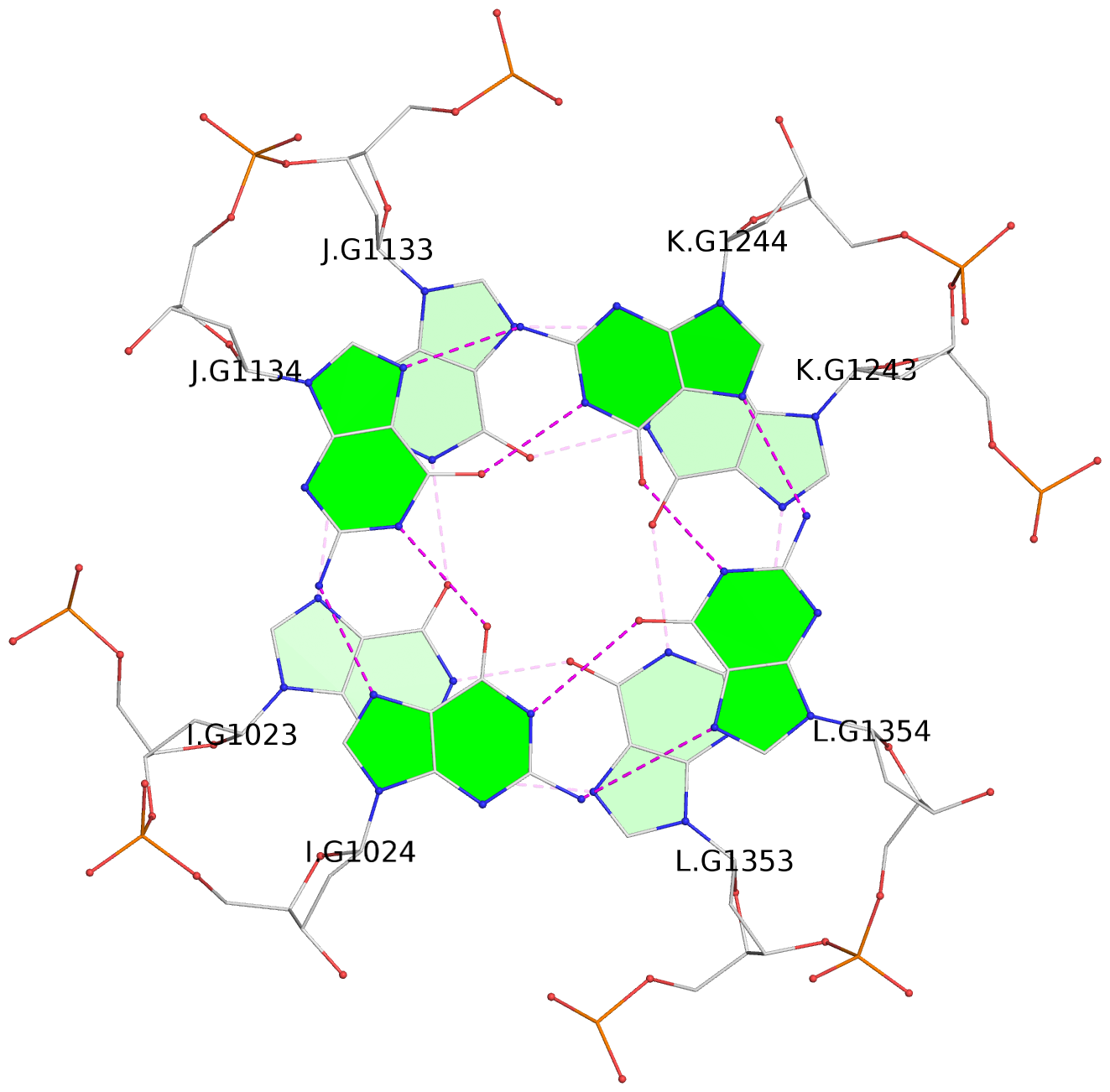
1 glyco-bond=---- sugar=---- groove=---- planarity=0.152 type=planar nts=4 GGGG A.DG102,D.DG432,C.DG322,B.DG212 2 glyco-bond=---- sugar=---- groove=---- planarity=0.186 type=bowl-2 nts=4 GGGG A.DG103,D.DG433,C.DG323,B.DG213 3 glyco-bond=---- sugar=.--- groove=---- planarity=0.173 type=other nts=4 GGGG A.DG104,D.DG434,C.DG324,B.DG214 4 glyco-bond=---- sugar=---- groove=---- planarity=0.280 type=bowl nts=4 GGGG A.DG105,D.DG435,C.DG325,B.DG215 5 glyco-bond=---- sugar=3333 groove=---- planarity=0.096 type=planar nts=4 GGGG E.DG582,H.DG912,G.DG802,F.DG692 6 glyco-bond=---- sugar=---- groove=---- planarity=0.236 type=other nts=4 GGGG E.DG583,H.DG913,G.DG803,F.DG693 7 glyco-bond=---- sugar=---- groove=---- planarity=0.165 type=other nts=4 GGGG E.DG584,H.DG914,G.DG804,F.DG694 8 glyco-bond=---- sugar=---- groove=---- planarity=0.245 type=bowl nts=4 GGGG E.DG585,H.DG915,G.DG805,F.DG695 9 glyco-bond=---- sugar=---- groove=---- planarity=0.331 type=bowl nts=4 GGGG I.DG1022,L.DG1352,K.DG1242,J.DG1132 10 glyco-bond=---- sugar=---- groove=---- planarity=0.246 type=bowl nts=4 GGGG I.DG1023,L.DG1353,K.DG1243,J.DG1133 11 glyco-bond=---- sugar=---- groove=---- planarity=0.242 type=other nts=4 GGGG I.DG1024,L.DG1354,K.DG1244,J.DG1134 12 glyco-bond=---- sugar=---- groove=---- planarity=0.237 type=other nts=4 GGGG I.DG1025,L.DG1355,K.DG1245,J.DG1135
List of 2 G4-helices
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 4 G-tetrad layers, inter-molecular, with 1 stem
Helix#2, 8 G-tetrad layers, inter-molecular, with 2 stems
List of 3 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 4 G-tetrad layers, 0 loops, inter-molecular, UUUU, parallel, parallel(4+0)
Stem#2, 4 G-tetrad layers, 0 loops, inter-molecular, UUUU, parallel, parallel(4+0)
Stem#3, 4 G-tetrad layers, 0 loops, inter-molecular, UUUU, parallel, parallel(4+0)
List of 1 G4 coaxial stack
1 G4 helix#2 contains 2 G4 stems: [#2,#3] [5'/5']