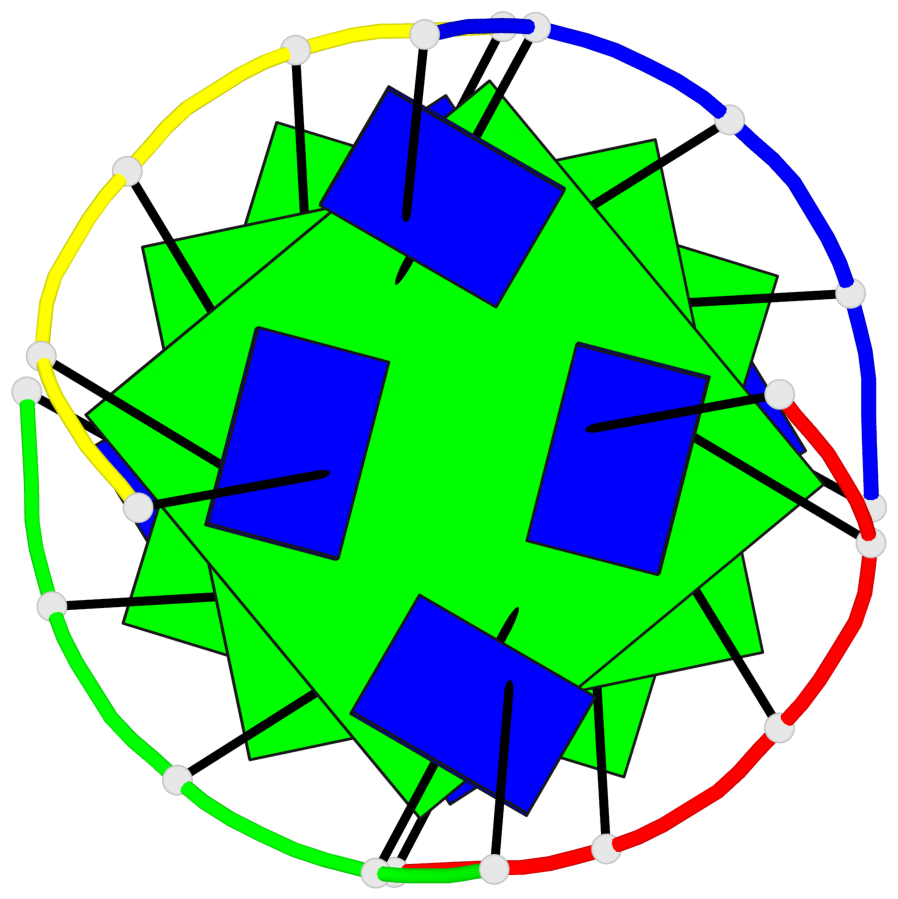
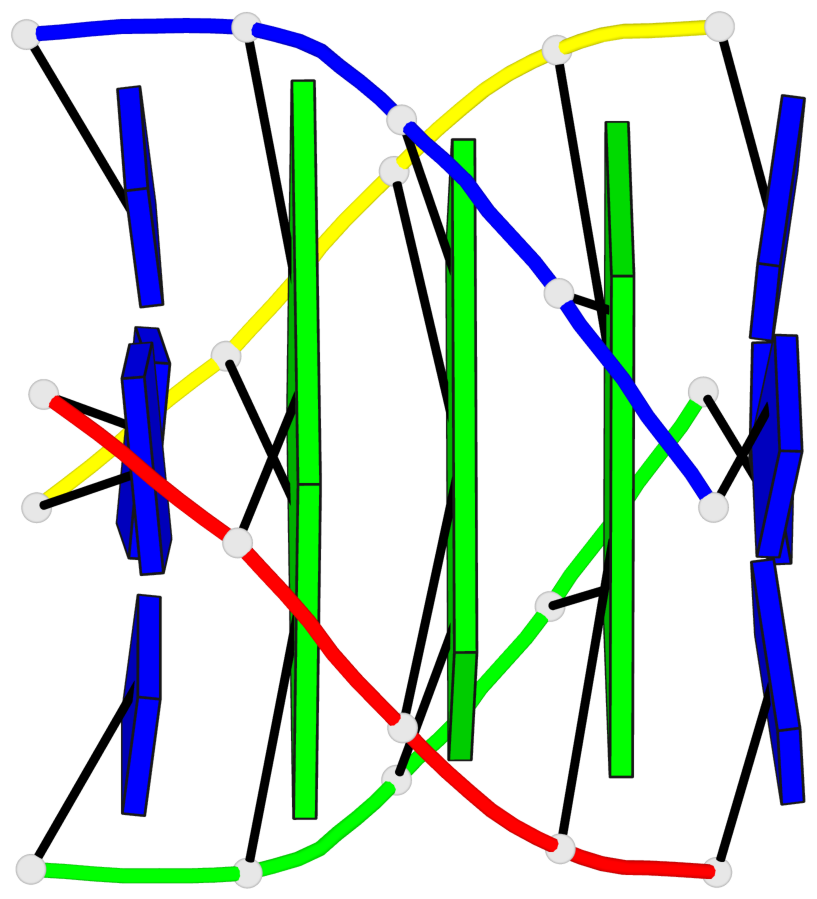
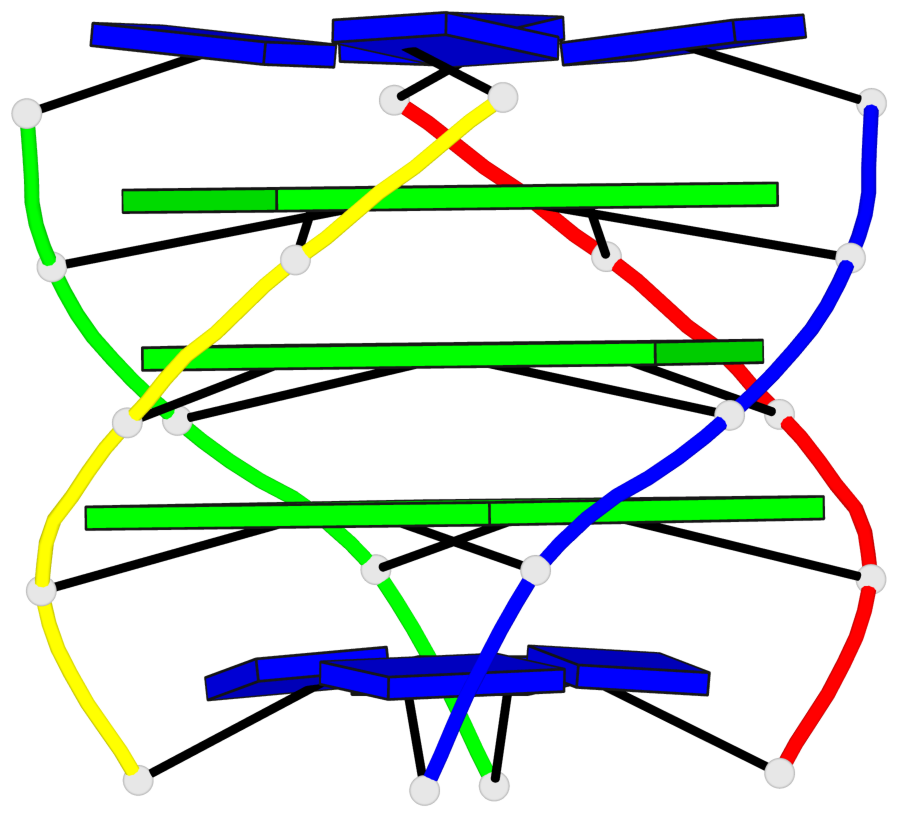
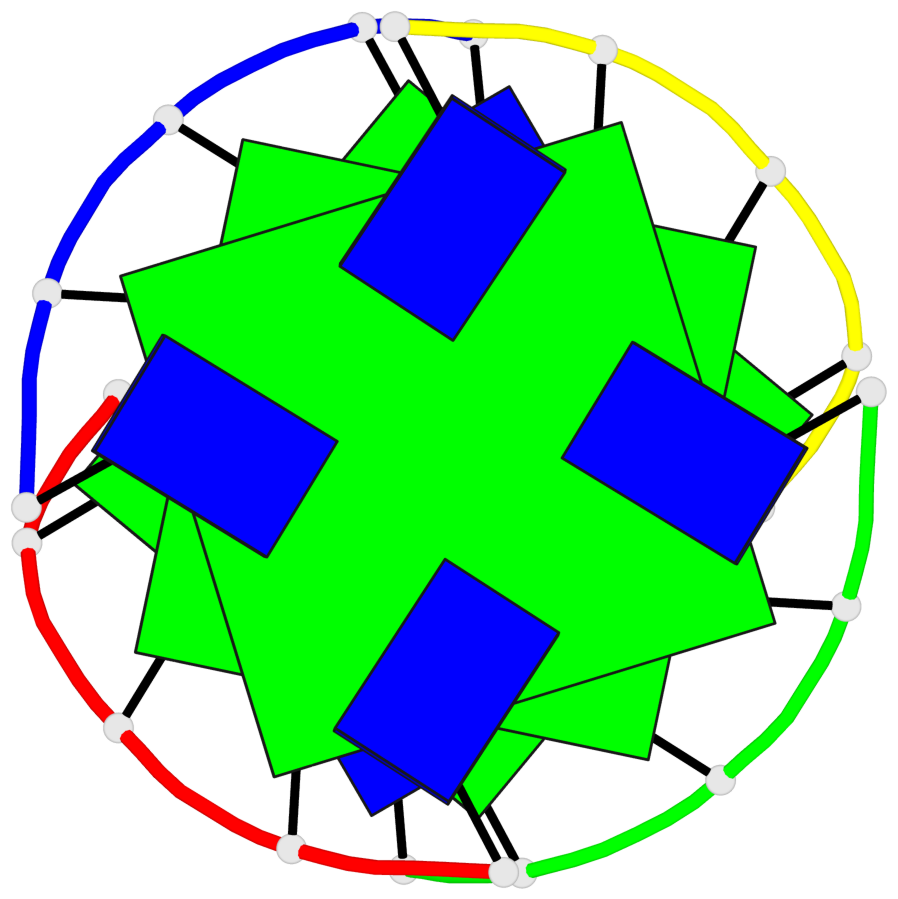
Detailed DSSR results for the G-quadruplex: PDB entry 1s9l
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 1s9l
- Class
- RNA
- Method
- NMR
- Summary
- NMR solution structure of a parallel lna quadruplex
- Reference
- Randazzo A, Esposito V, Ohlenschlager O, Ramachandran R, Mayola L (2004): "NMR solution structure of a parallel LNA quadruplex." Nucleic Acids Res., 32, 3083-3092. doi: 10.1093/nar/gkh629.
- Abstract
- The solution structure of a locked nucleic acid (LNA) quadruplex, formed by the oligomer d(TGGGT), containing only conformationally restricted LNA residues is reported. NMR and CD spectroscopy, as well as molecular dynamics and mechanic calculations, has been used to characterize the complex. The molecule adopts a parallel stranded conformation with a 4-fold rotational symmetry, showing a right-handed helicity and the guanine residues in an almost planar conformation with three well-defined G-tetrads. The thermal stability of Q-LNA has been found to be comparable with that of [r(UGGGU)]4, while a T(m) increment of 20 degrees C with respect to the corresponding DNA quadruplex structure [d(TGGGT)]4 has been observed. The structural features of the LNA quadruplex reported here may open new perspectives for the biological application of LNAs as novel versatile tools to design aptamer or catalyst oligonucleotides.
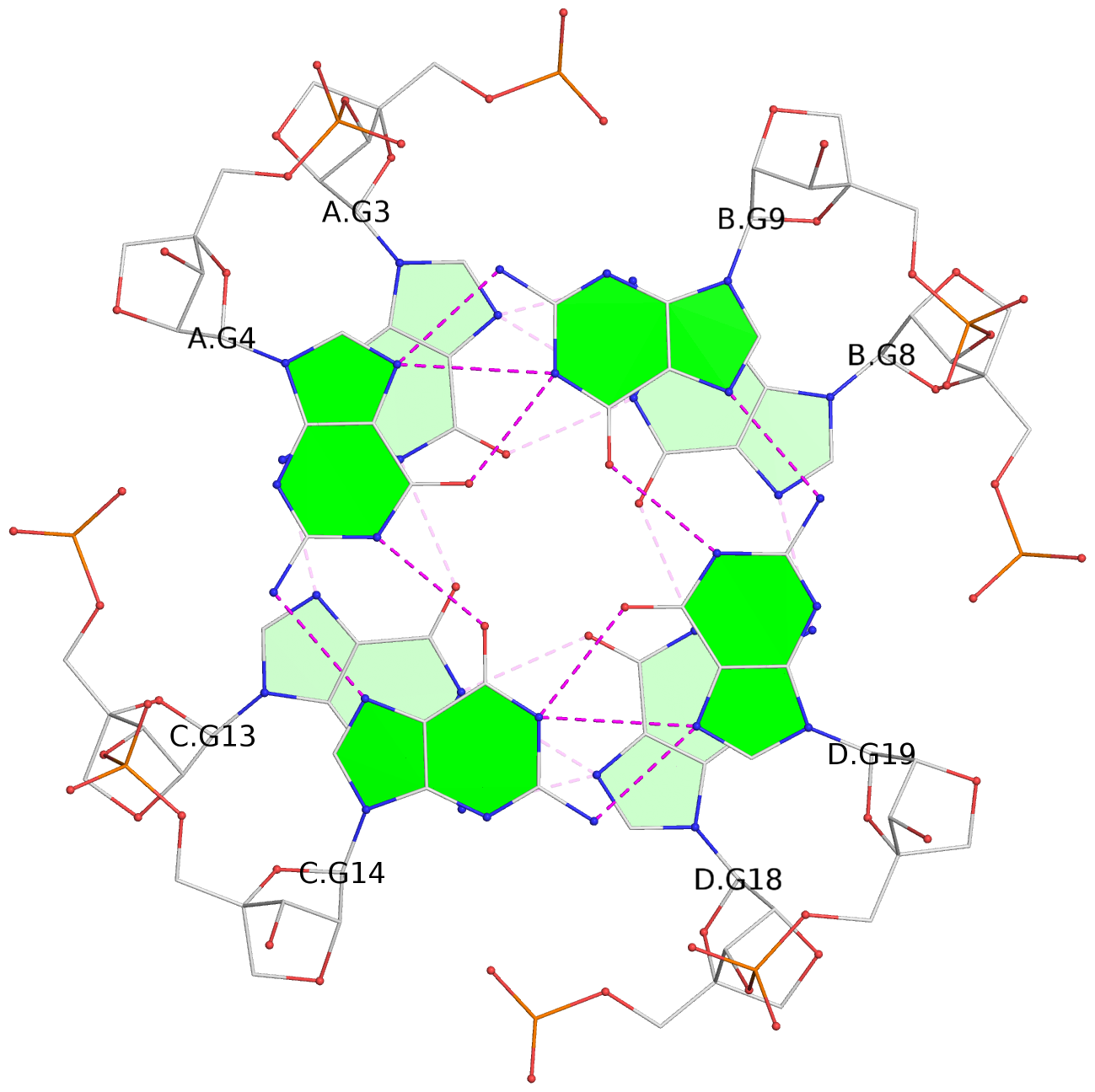
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, parallel(4+0), UUUU
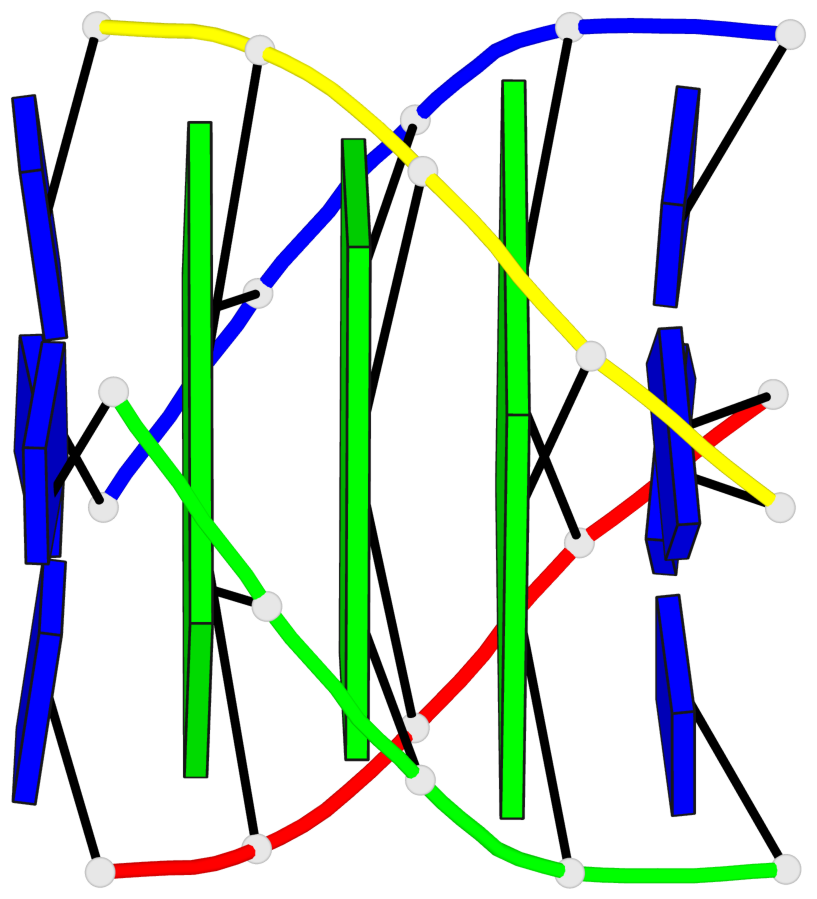
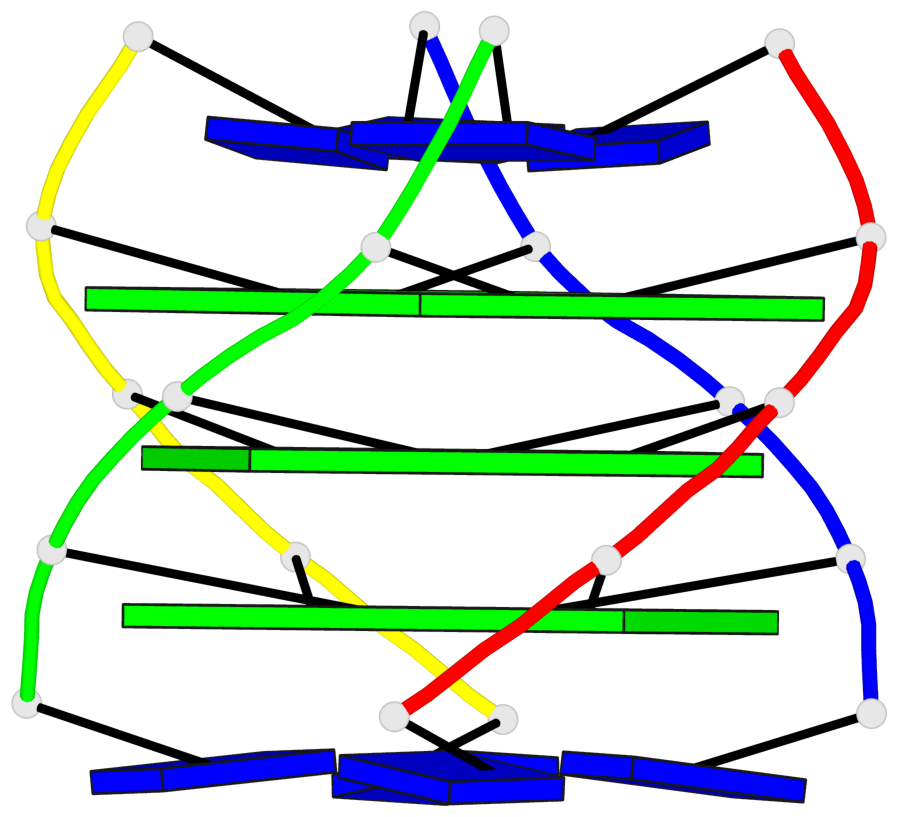
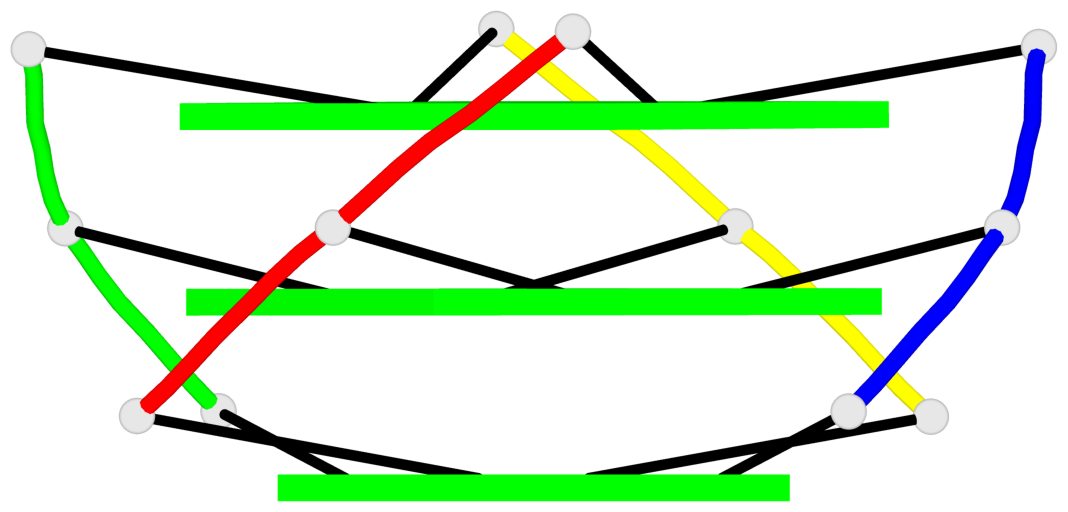
Base-block schematics in six views
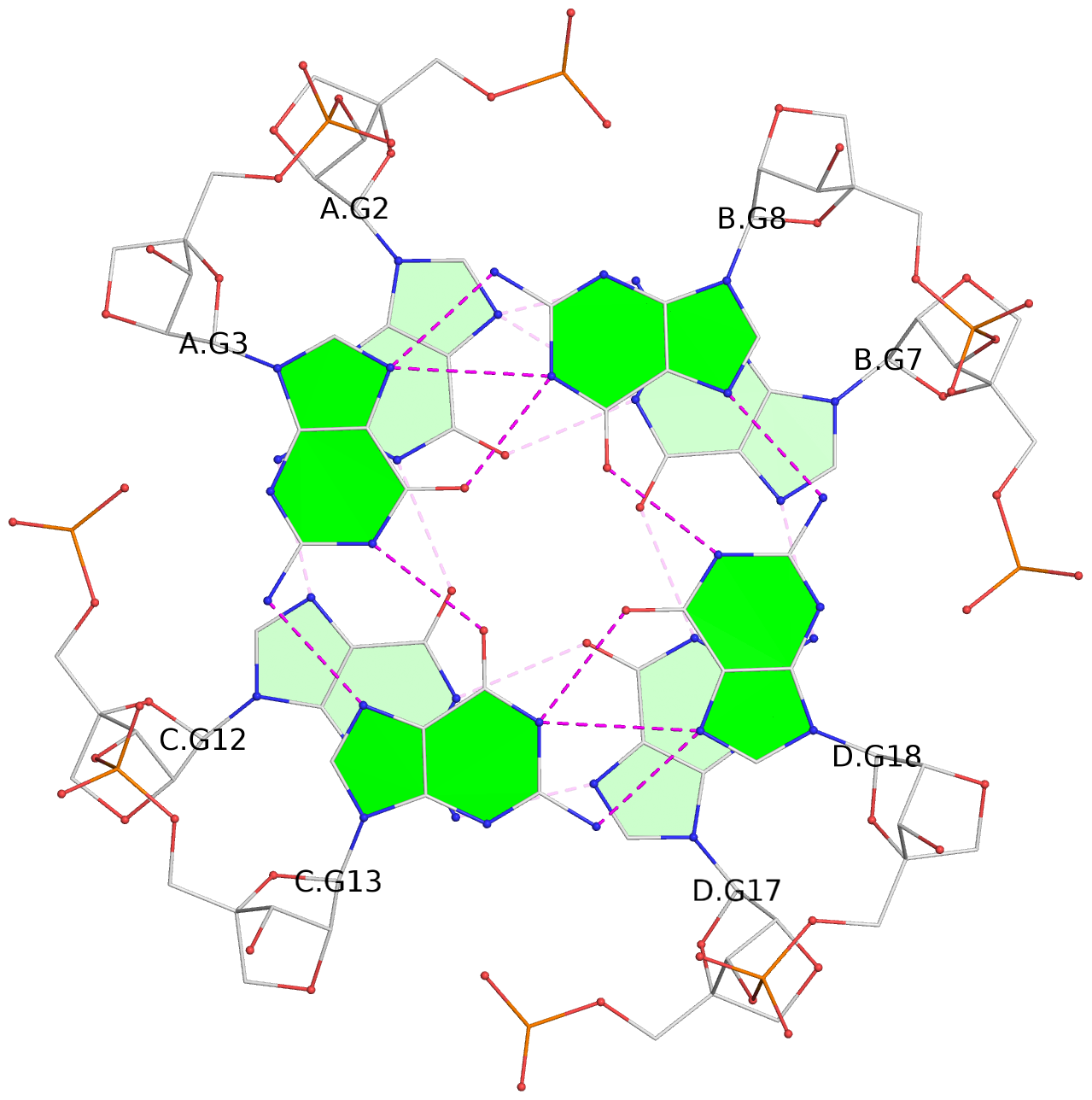
List of 3 G-tetrads
1 glyco-bond=---- sugar=3333 groove=---- planarity=0.227 type=other nts=4 gggg A.LCG2,C.LCG12,D.LCG17,B.LCG7 2 glyco-bond=---- sugar=3333 groove=---- planarity=0.208 type=other nts=4 gggg A.LCG3,C.LCG13,D.LCG18,B.LCG8 3 glyco-bond=---- sugar=3333 groove=---- planarity=0.191 type=other nts=4 gggg A.LCG4,C.LCG14,D.LCG19,B.LCG9
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, inter-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.