Detailed DSSR results for the G-quadruplex: PDB entry 1u64
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 1u64
- Class
- DNA
- Method
- NMR
- Summary
- The solution structure of d(g3t4g4)2
- Reference
- Sket P, Crnugelj M, Plavec J (2004): "d(G3T4G4) forms unusual dimeric G-quadruplex structure with the same general fold in the presence of K+, Na+ or NH4+ ions." Bioorg.Med.Chem., 12, 5735-5744. doi: 10.1016/j.bmc.2004.08.009.
- Abstract
- We have recently communicated that DNA oligonucleotide d(G(3)T(4)G(4)) forms a dimeric G-quadruplex in the presence of K(+) ions [J. Am. Chem. Soc.2003, 125, 7866-7871]. The high-resolution NMR structure of d(G(3)T(4)G(4))(2) G-quadruplex exhibits G-quadruplex core consisting of three stacked G-quartets. The two overhanging G3 and G11 residues are located at the opposite sides of the end G-quartets and are not involved in G-quartet formation. d(G(3)T(4)G(4))(2) G-quadruplex represents the first bimolecular G-quadruplex where end G-quartets are spanned by diagonal (T4-T7) as well as edge-type loops (T15-T18). Three of the G-rich strands are parallel while one is anti-parallel. The G12-G22 strand demonstrates a sharp reversal in strand direction between residues G19 and G20 that is accommodated with the leap over the middle G-quartet. The reversal in strand direction is achieved without any extra intervening residues. Here we furthermore examined the influence of different monovalent cations on the folding of d(G(3)T(4)G(4)). The resolved imino and aromatic proton resonances as well as (sequential) NOE connectivity patterns showed only minor differences in key intra- and interquartet NOE intensities in the presence of K(+), Na(+) and NH(4)(+) ions, which were consistent with subtle structural differences while retaining the same folding topology of d(G(3)T(4)G(4))(2) G-quadruplex.
- G4 notes
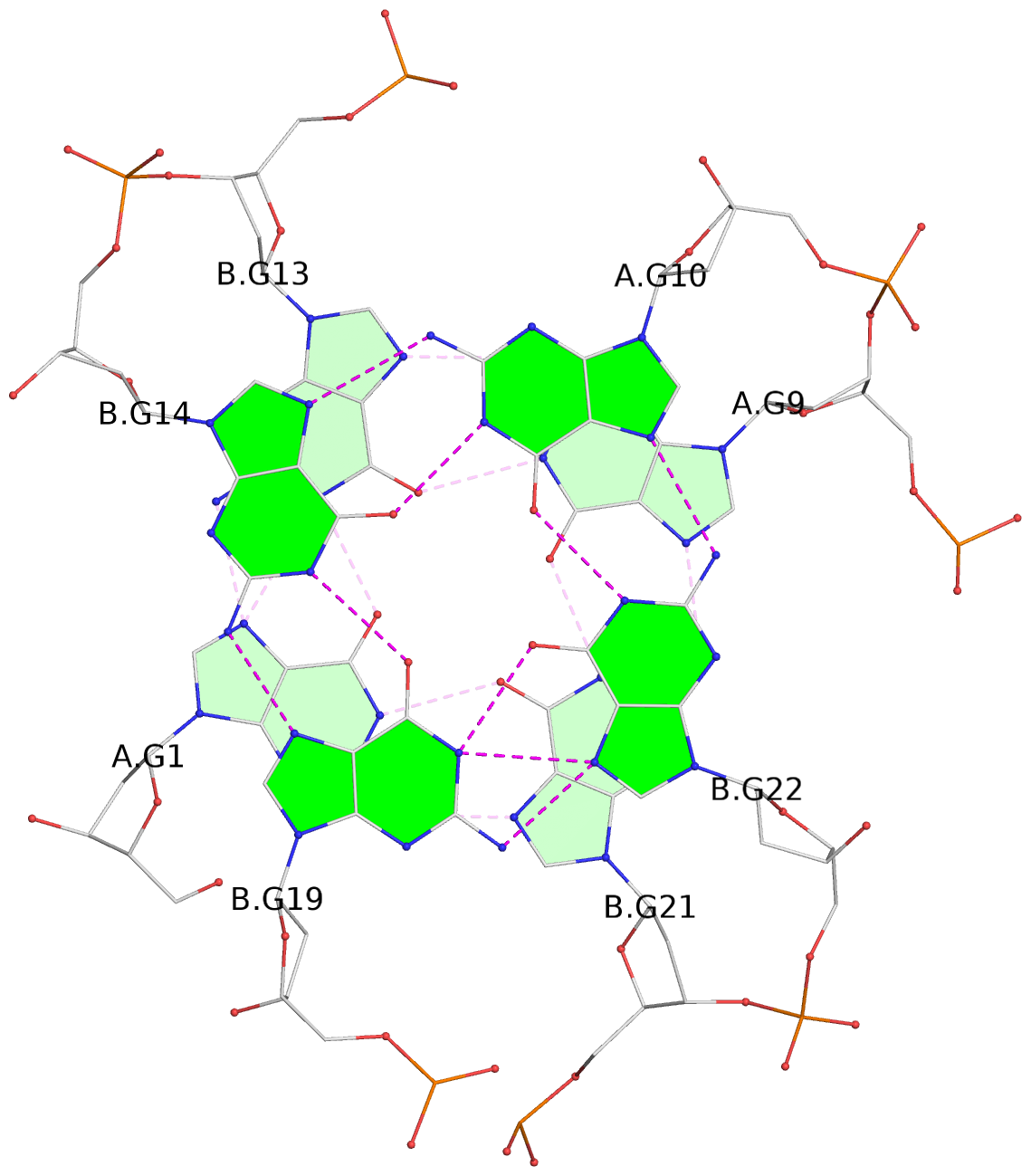
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, (1+3), UDDD
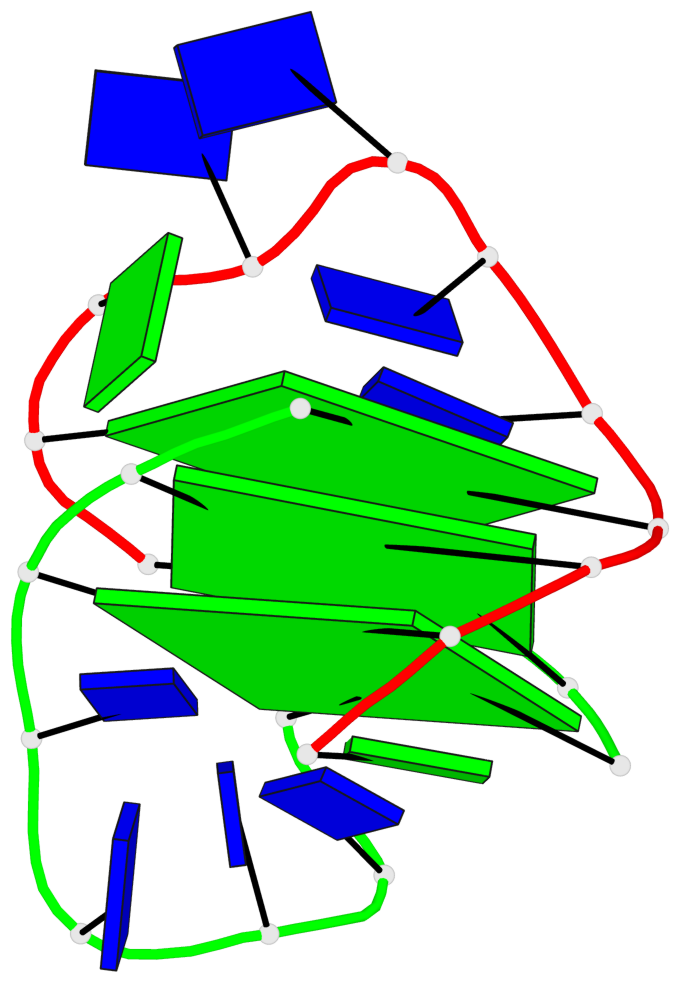
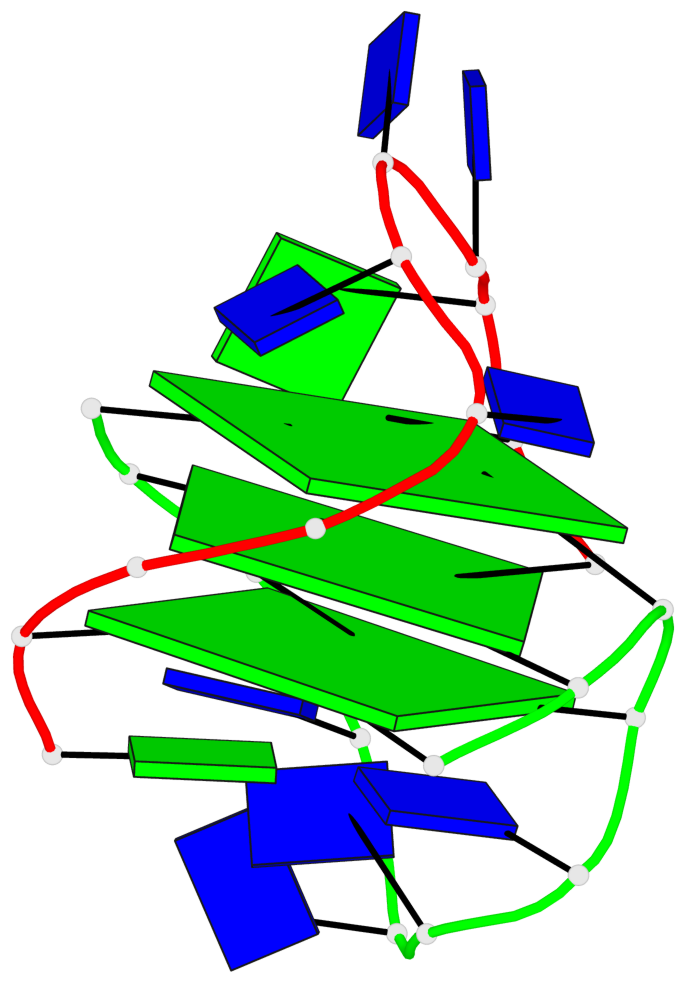
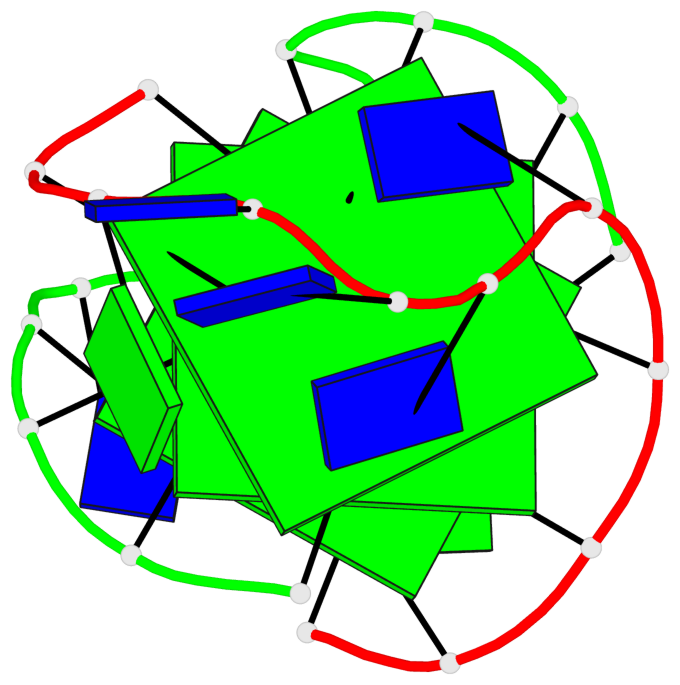
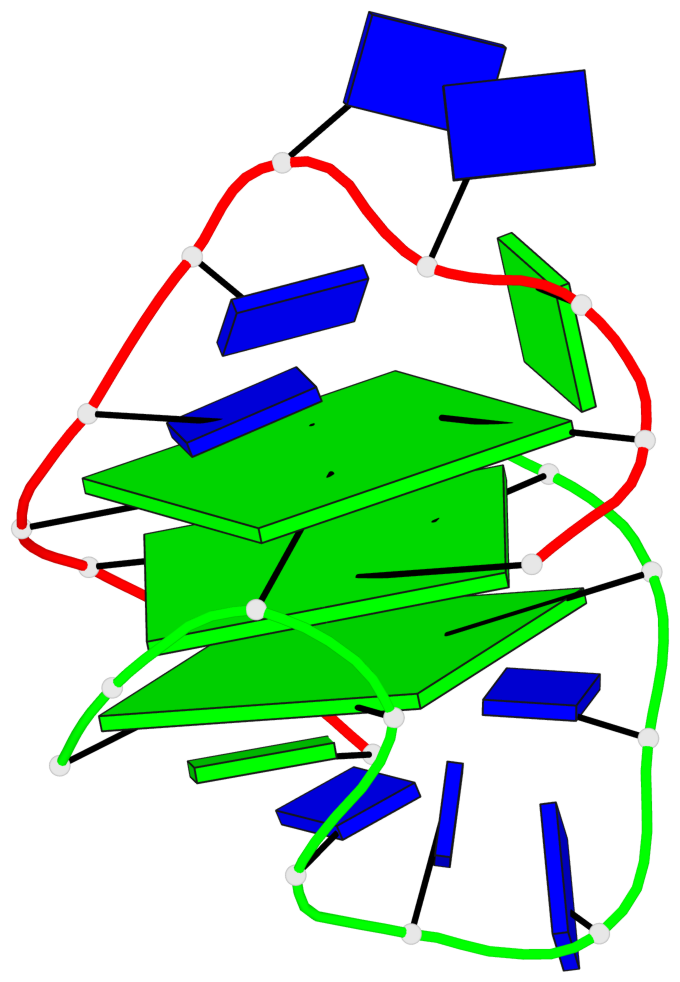
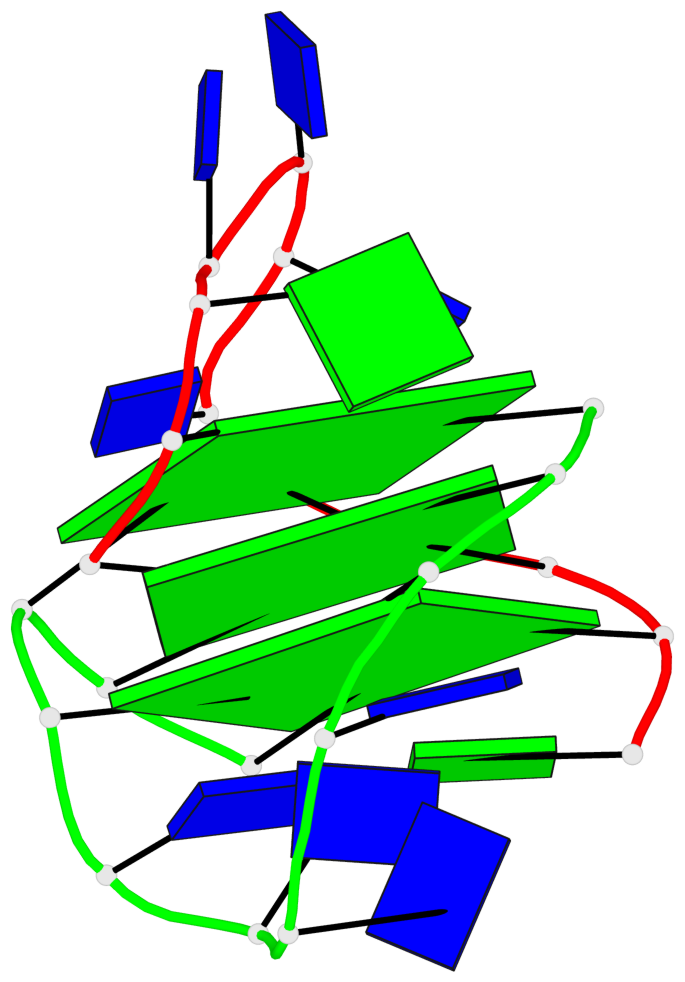
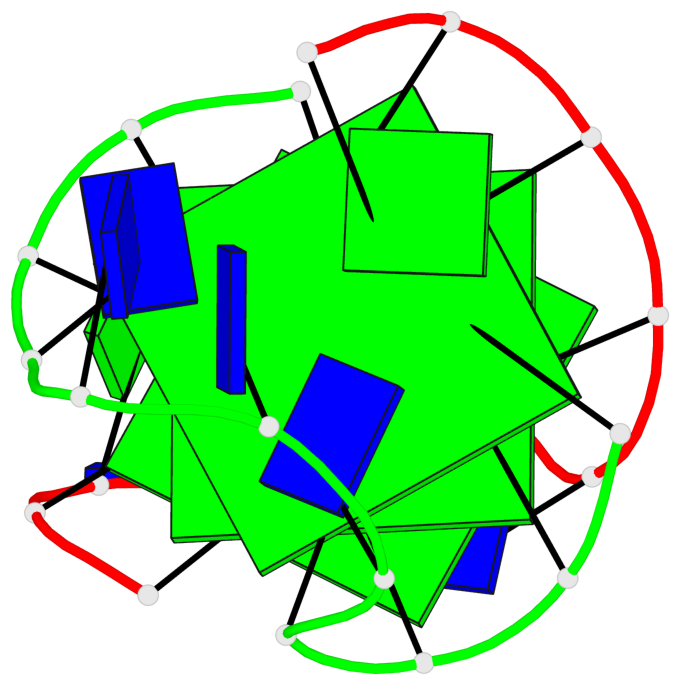
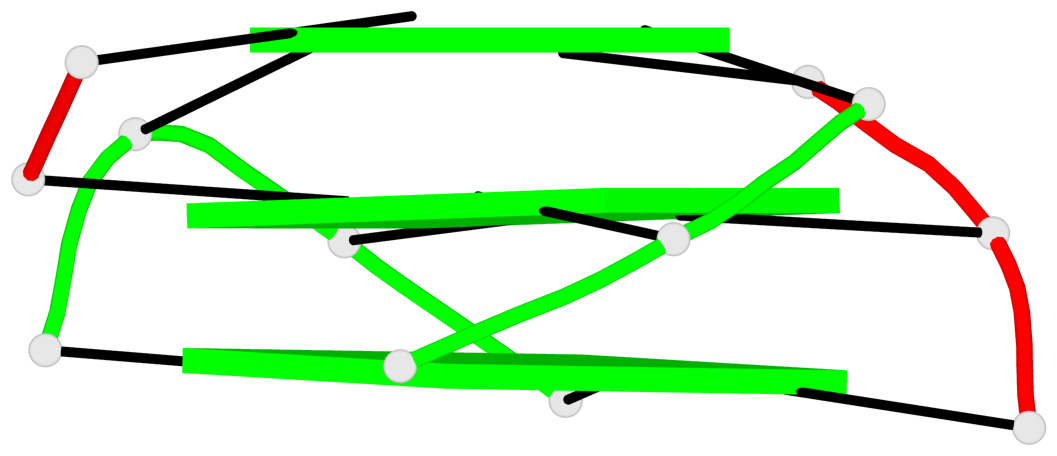
Base-block schematics in six views
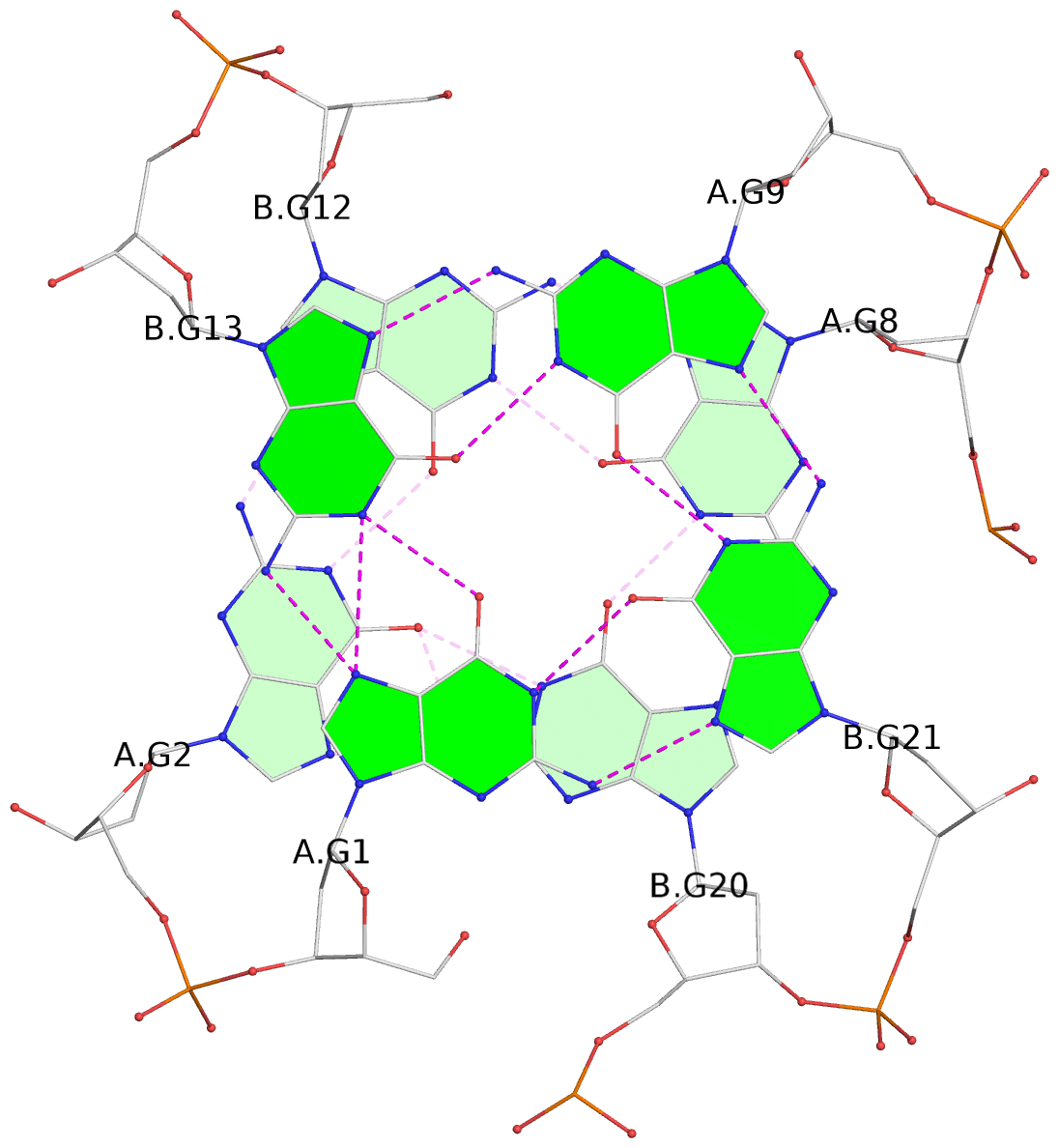
List of 3 G-tetrads
1 glyco-bond=s--- sugar=---- groove=w--n planarity=0.178 type=other nts=4 GGGG A.DG1,B.DG13,A.DG9,B.DG21 2 glyco-bond=-ss- sugar=---- groove=w-n- planarity=0.240 type=other nts=4 GGGG A.DG2,B.DG12,A.DG8,B.DG20 3 glyco-bond=--s- sugar=---- groove=-wn- planarity=0.263 type=other nts=4 GGGG A.DG10,B.DG14,B.DG19,B.DG22
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, inter-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 2 G-tetrad layers, 2 loops, inter-molecular, UDDD, hybrid-(mixed), (1+3)
List of 2 non-stem G4-loops (including the two closing Gs)
1 type=lateral helix=#1 nts=6 GTTTTG B.DG14,B.DT15,B.DT16,B.DT17,B.DT18,B.DG19 2 type=V-shaped helix=#1 nts=4 GGGG B.DG19,B.DG20,B.DG21,B.DG22