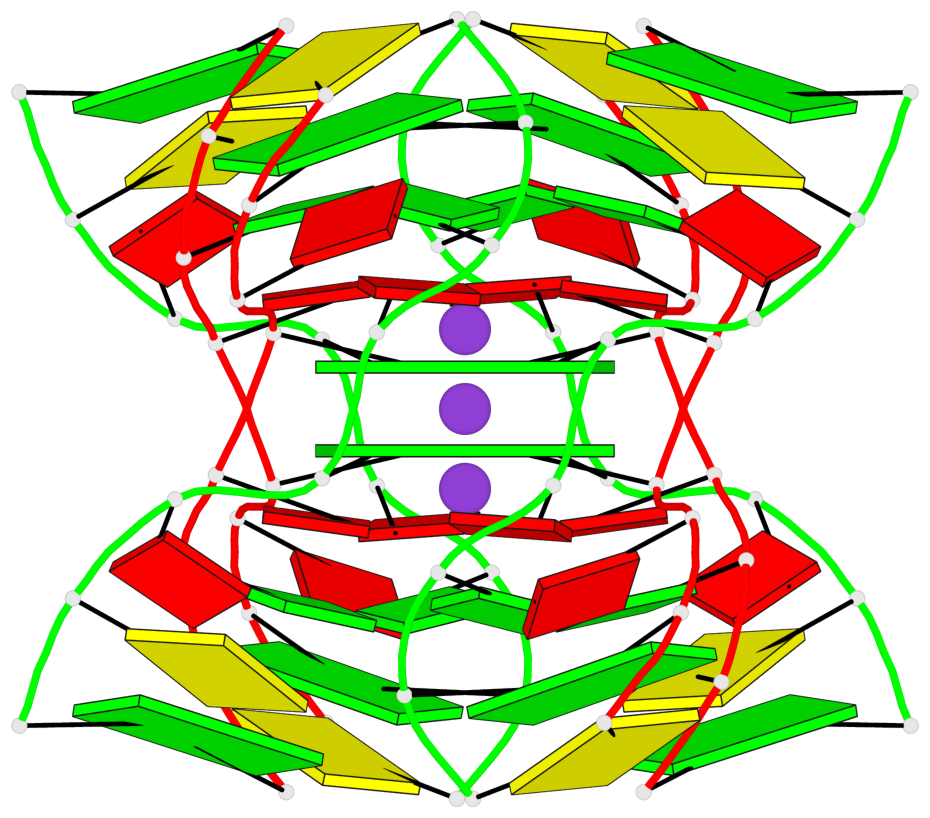
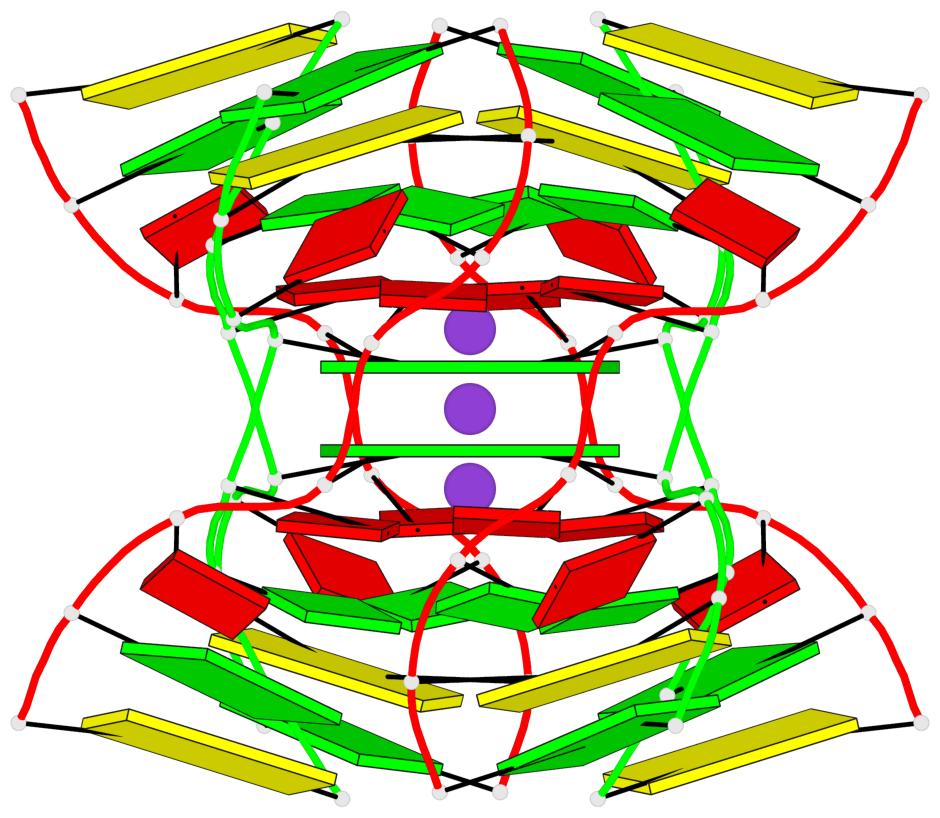
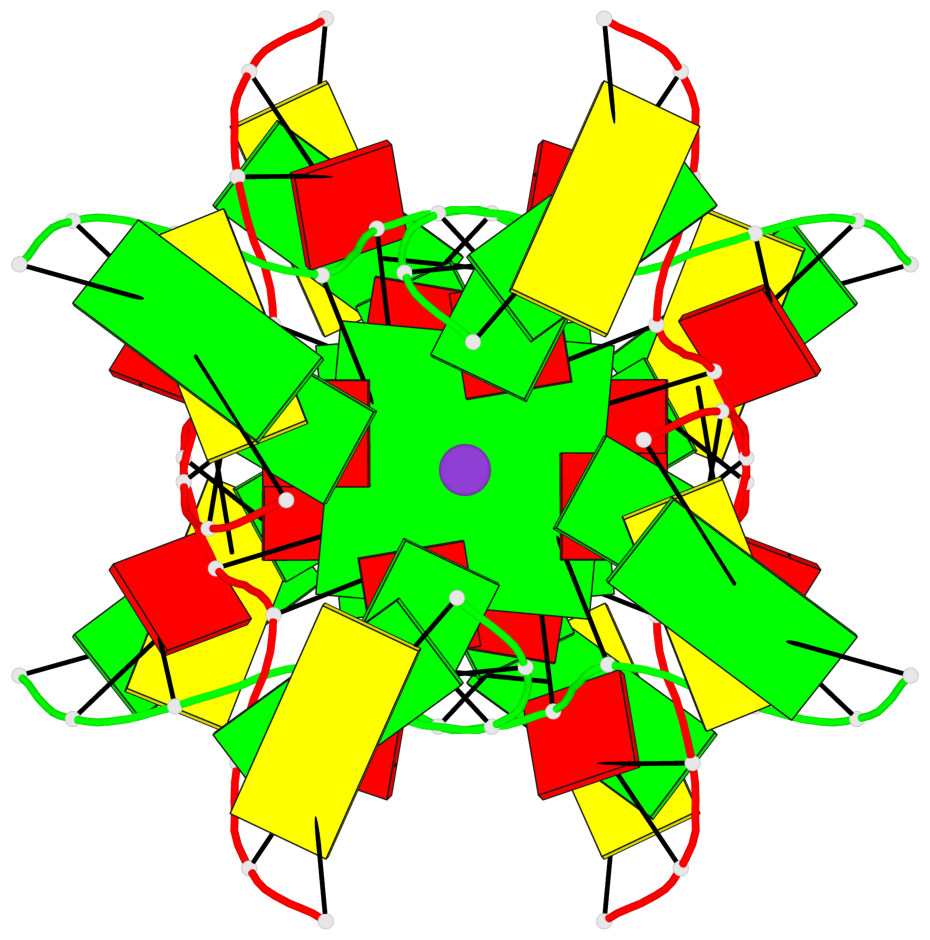
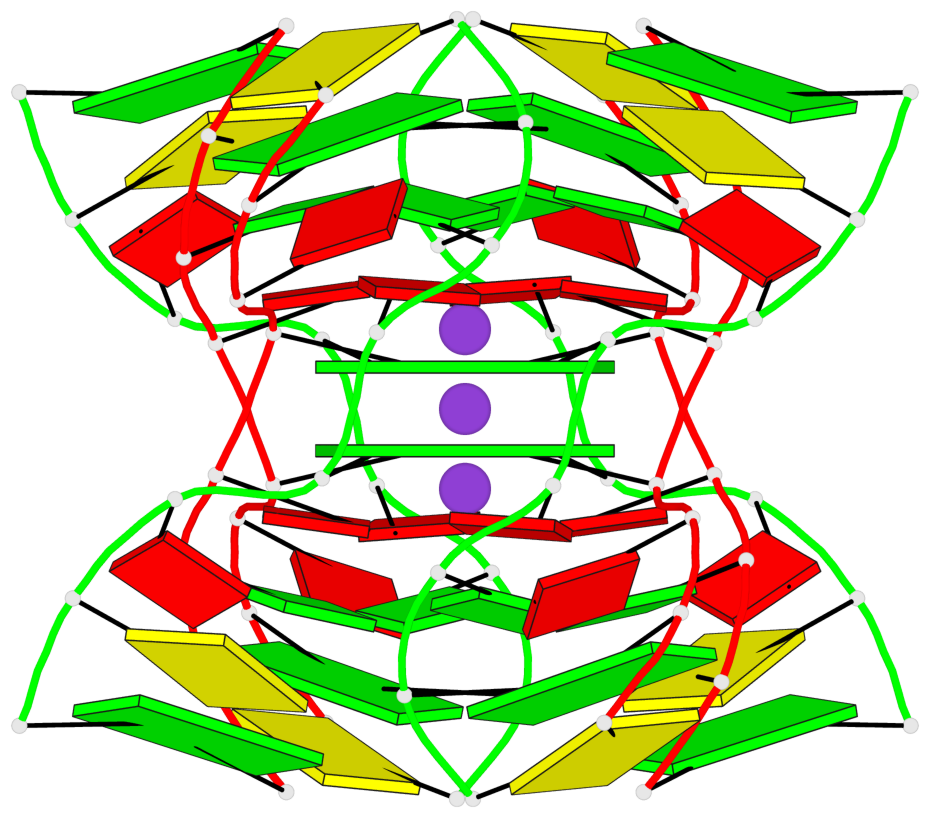
Detailed DSSR results for the G-quadruplex: PDB entry 1v3p
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 1v3p
- Class
- DNA
- Method
- X-ray (2.3 Å)
- Summary
- Crystal structure of d(gcgagagc): the DNA octaplex structure with i-motif of g-quartet
- Reference
- Kondo J, Adachi W, Umeda S, Sunami T, Takenaka A (2004): "Crystal structures of a DNA octaplex with I-motif of G-quartets and its splitting into two quadruplexes suggest a folding mechanism of eight tandem repeats." Nucleic Acids Res., 32, 2541-2549. doi: 10.1093/nar/gkh575.
- Abstract
- Recent genomic analyses revealed many kinds of tandem repeats of specific sequences. Some of them are related to genetic diseases, but their biological functions and structures are still unknown. Two X-ray structures of a short DNA fragment d(gcGA[G]1Agc) show that four base-intercalated duplexes are assembled to form an octaplex at a low K+ concentration, in which the eight G5 residues form a stacked double G-quartet in the central part. At a higher K+ concentration, however, the octaplex is split into just two halves. These structural features suggest a folding process of eight tandem repeats of d(ccGA[G]4Agg), according to a double Greek-key motif. Such a packaging of the repeats could facilitate slippage of a certain sequence during DNA replication, to induce increase or decrease of the repeats.
- G4 notes
- 2 G-tetrads, 1 G4 helix
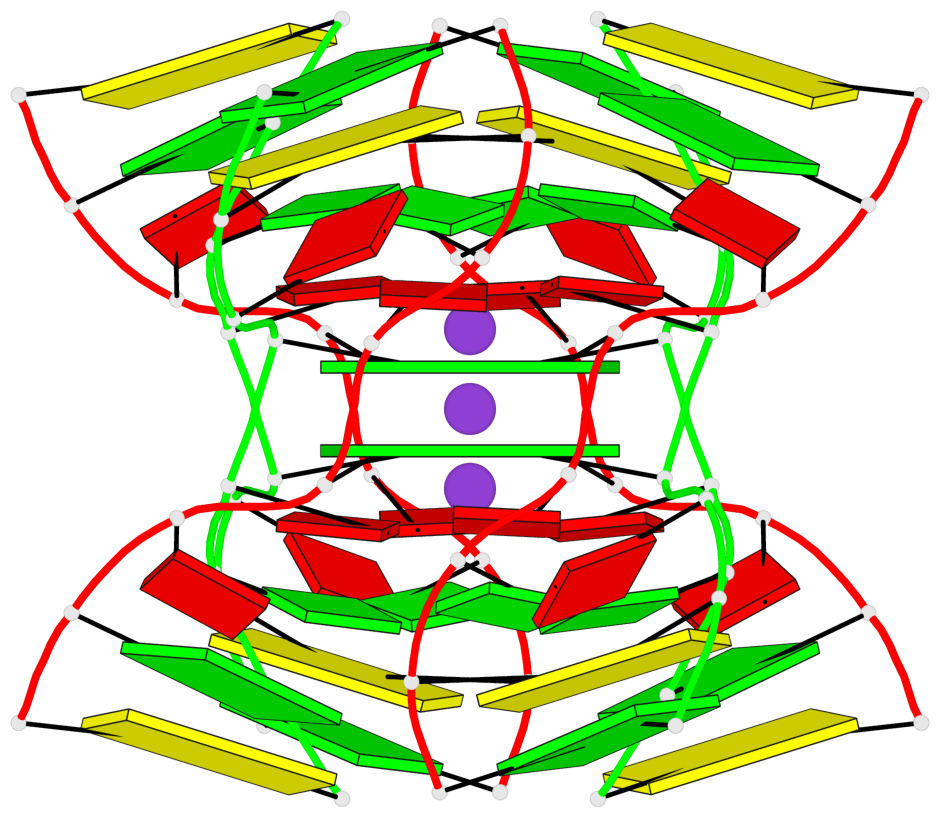
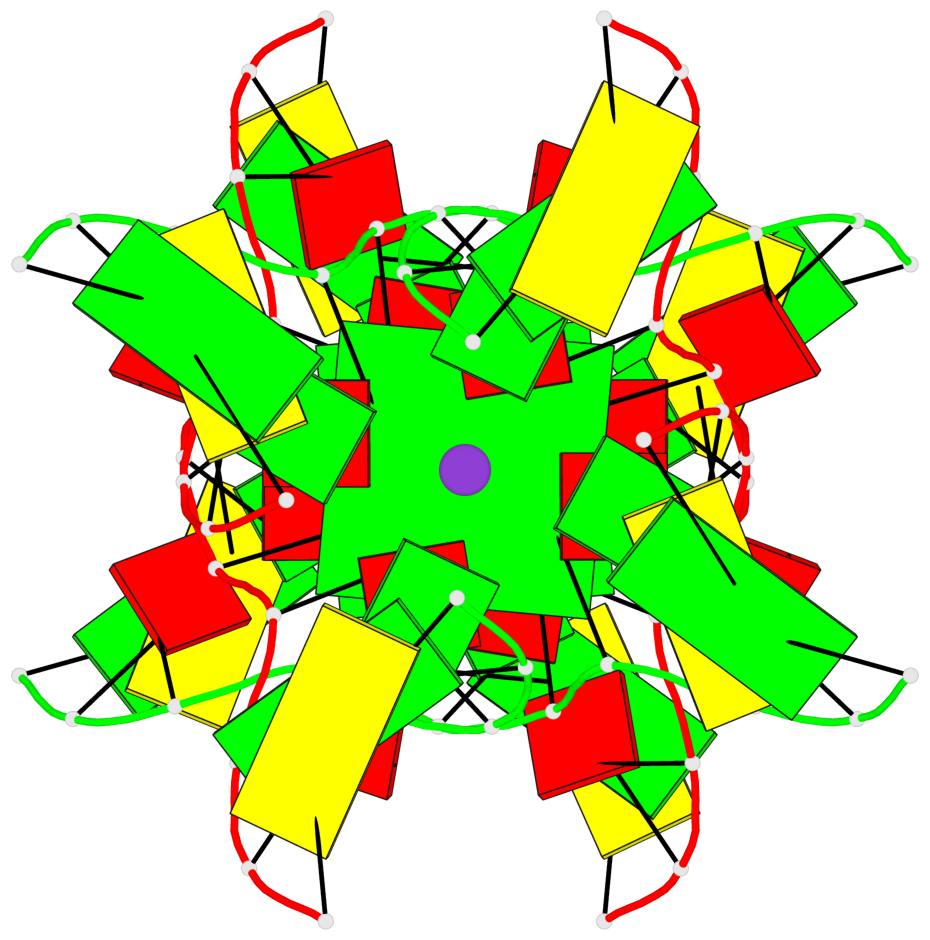
Base-block schematics in six views
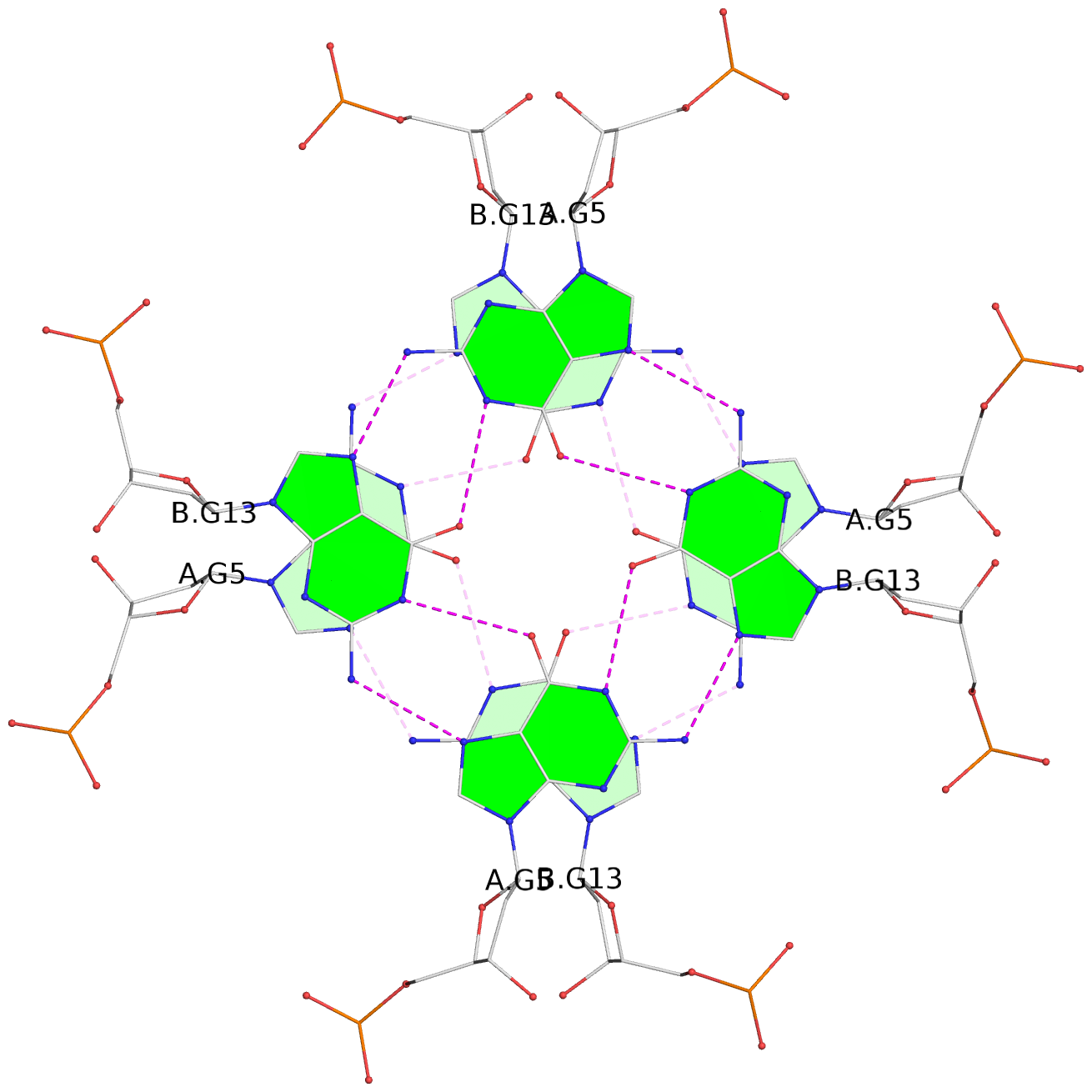
List of 2 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.115 type=planar nts=4 GGGG 1:A.DG5,3:B.DG13,2:A.DG5,4:B.DG13 2 glyco-bond=---- sugar=---- groove=---- planarity=0.115 type=planar nts=4 GGGG 1:B.DG13,4:A.DG5,2:B.DG13,3:A.DG5
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.