Detailed DSSR results for the G-quadruplex: PDB entry 1y8d
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 1y8d
- Class
- DNA
- Method
- NMR
- Summary
- Dimeric parallel-stranded tetraplex with 3+1 5' g-tetrad interface, single-residue chain reversal loops and gag triad in the context of a(gggg) pentad
- Reference
- Phan AT, Kuryavyi VV, Ma J-B, Faure A, Andreola M-L, Patel DJ (2005): "An interlocked dimeric parallel-stranded DNA quadruplex: A potent inhibitor of HIV-1 integrase." Proc.Natl.Acad.Sci.USA, 102, 634-639. doi: 10.1073/pnas.0406278102.
- Abstract
- We report on the NMR-based solution structure of the 93del d(GGGGTGGGAGGAGGGT) aptamer, a potent nanomolar inhibitor of HIV-1 integrase. This guanine-rich DNA sequence adopts an unusually stable dimeric quadruplex architecture in K+ solution. Within each 16-nt monomer subunit, which contains one A.(G.G.G.G) pentad sandwiched between two G.G.G.G tetrads, all G-stretches are parallel, and all guanines are anti with the exception of G1, which is syn. Dimer formation is achieved through mutual pairing of G1 of one monomer, with G2, G6, and G13 of the other monomer, to complete G.G.G.G tetrad formation. There are three single-nucleotide double-chain-reversal loops within each monomer fold, such that the first (T5) and third (A12) loops bridge three G-tetrad layers, whereas the second (A9) loop bridges two G-tetrad layers and participates in A.(G.G.G.G) pentad formation. Results of NMR and of integrase inhibition assays on loop-modified sequences allowed us to propose a strategy toward the potential design of improved HIV-1 integrase inhibitors. Finally, we propose a model, based on molecular docking approaches, for positioning the 93del dimeric DNA quadruplex within a basic channel/canyon formed between subunits of a dimer of dimers of HIV-1 integrase.
- G4 notes
- 6 G-tetrads, 1 G4 helix, 2 G4 stems, 1 G4 coaxial stack, 2(-P-P-P), parallel(4+0), UUUU, coaxial interfaces: 5'/5'-SEPARATED
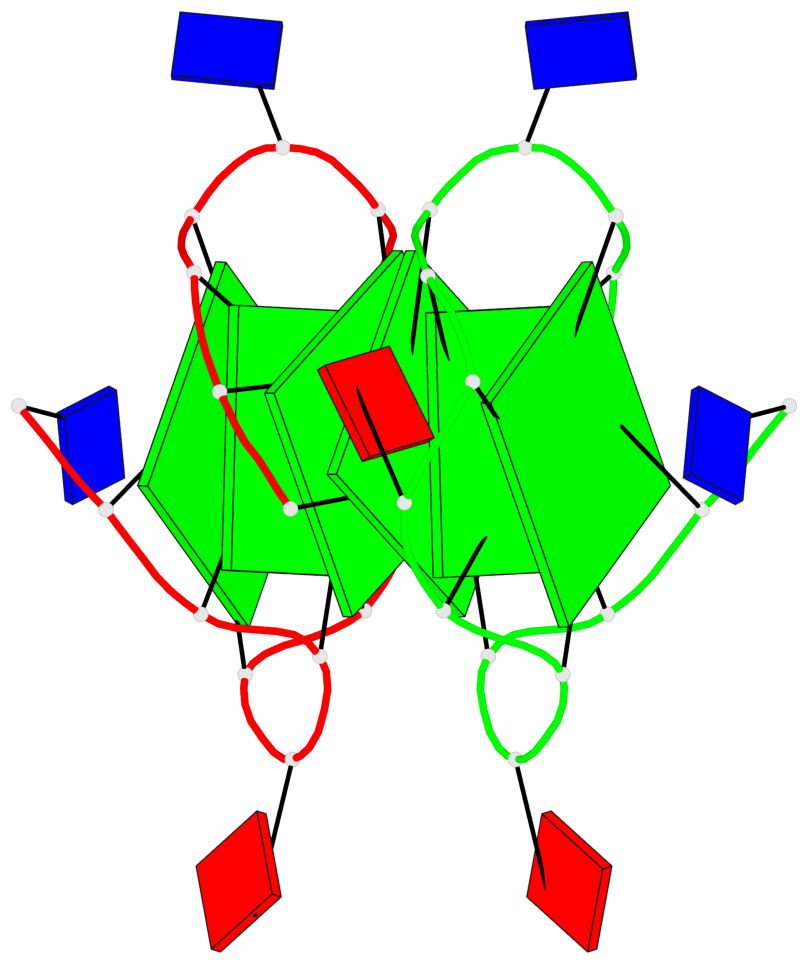
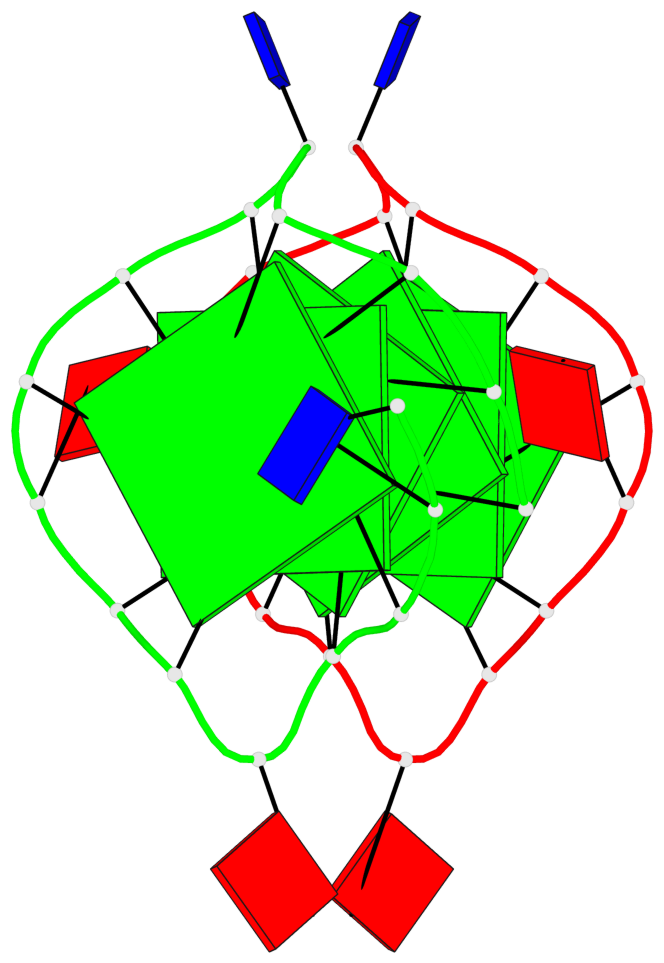
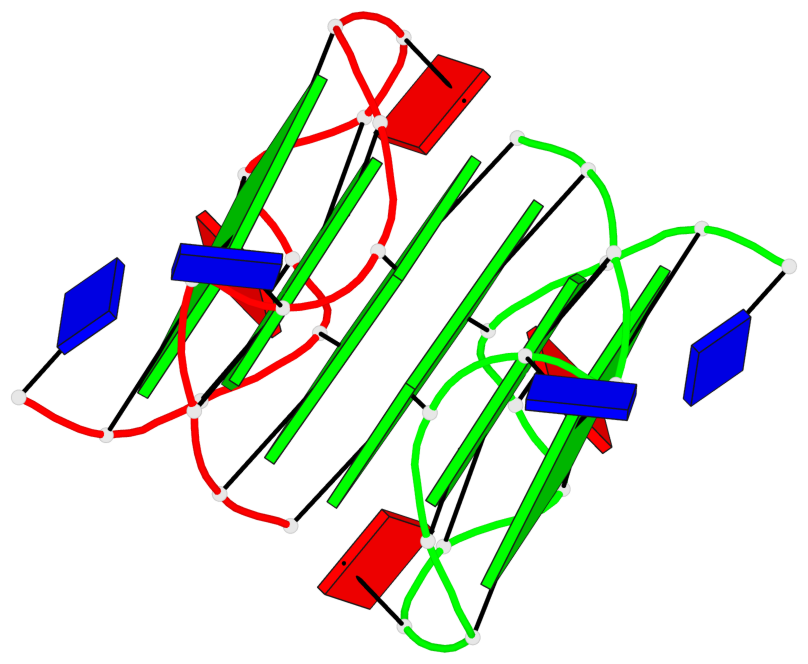
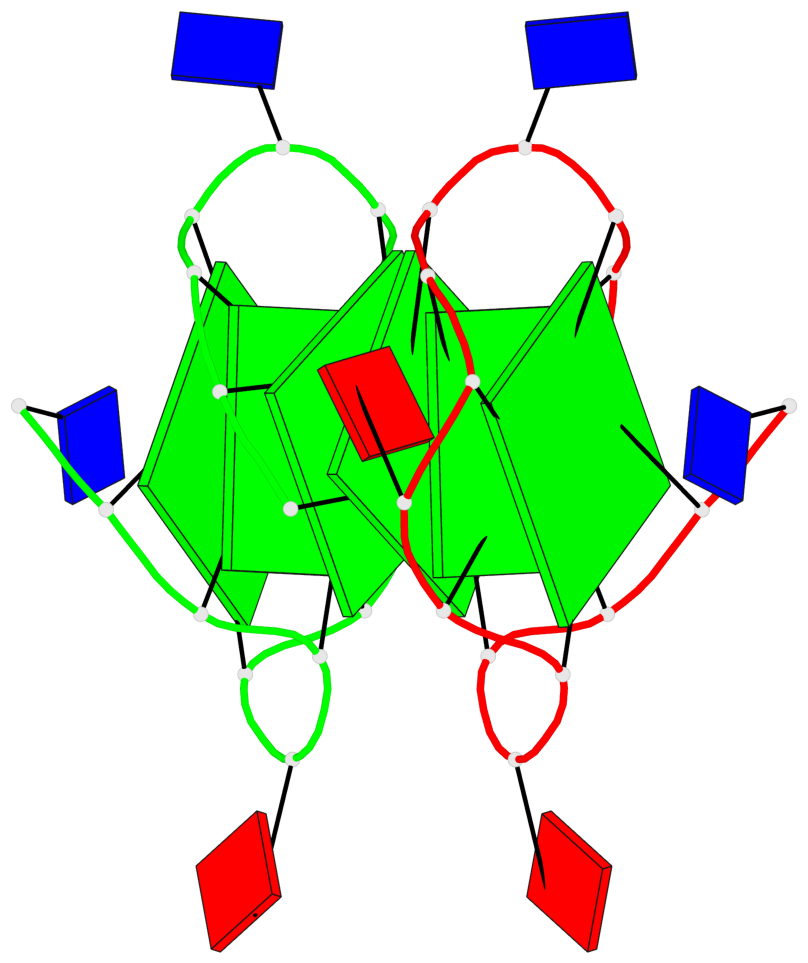
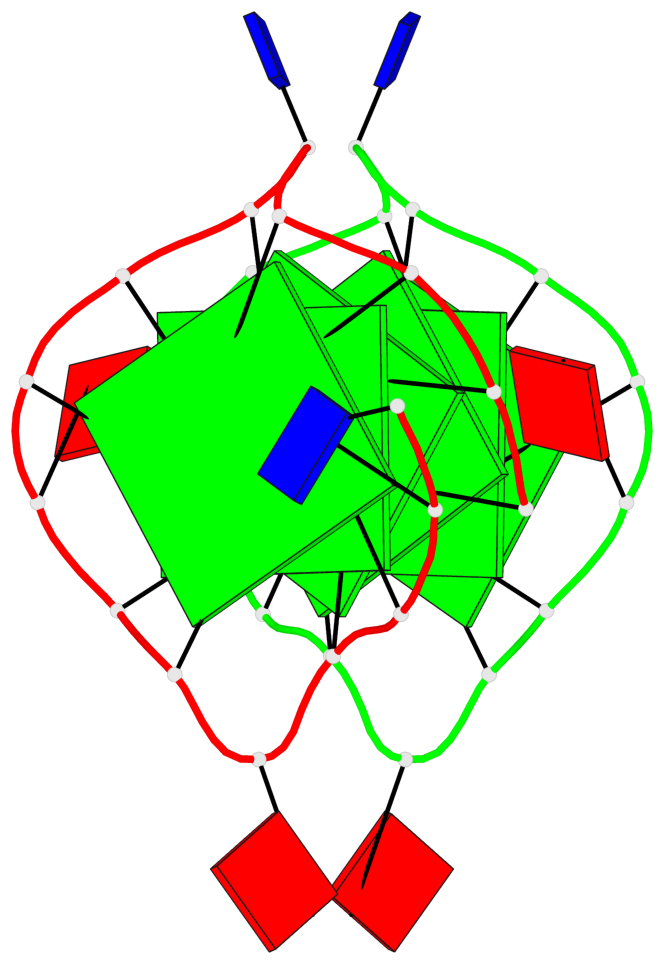
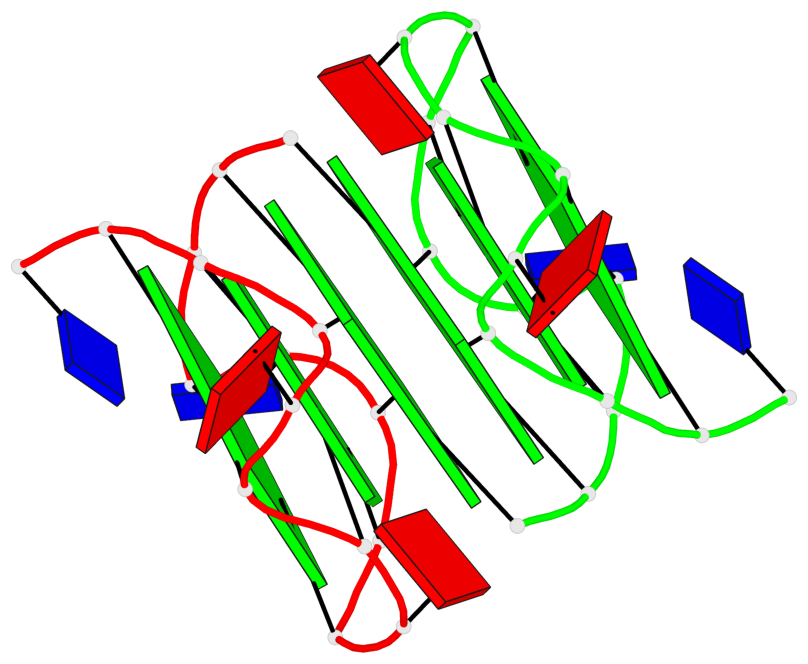
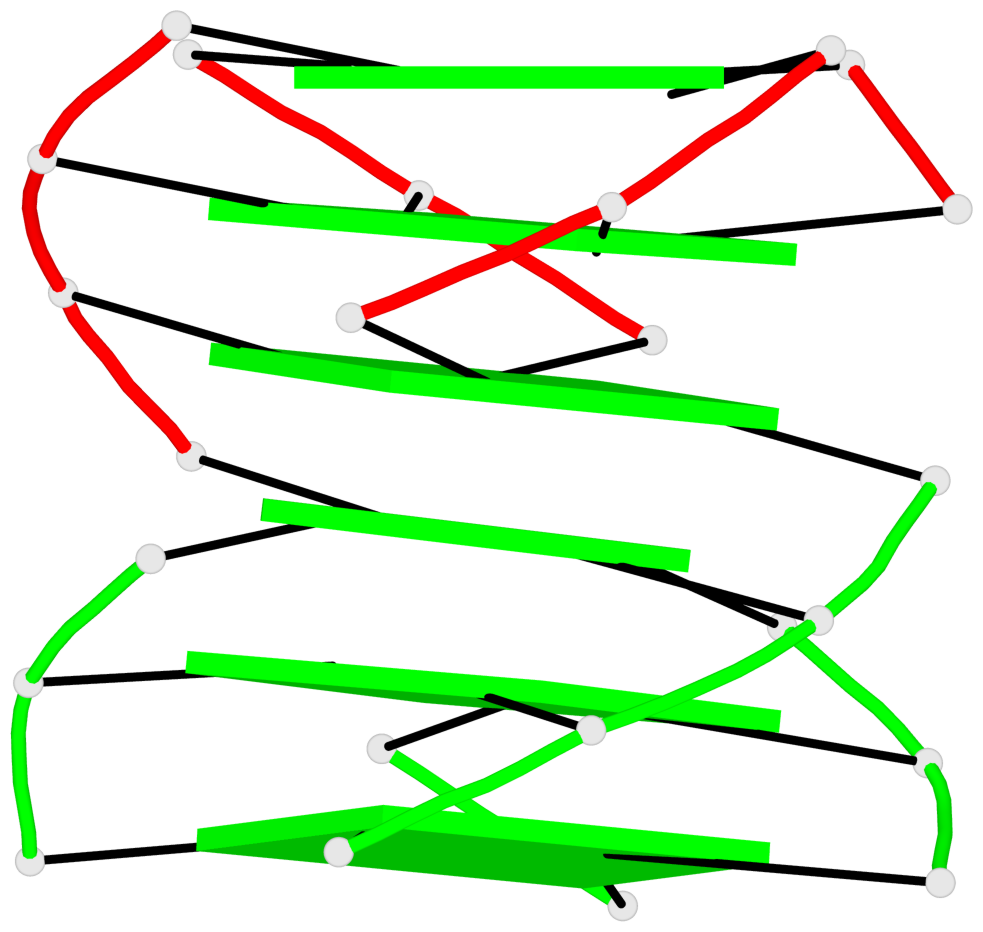
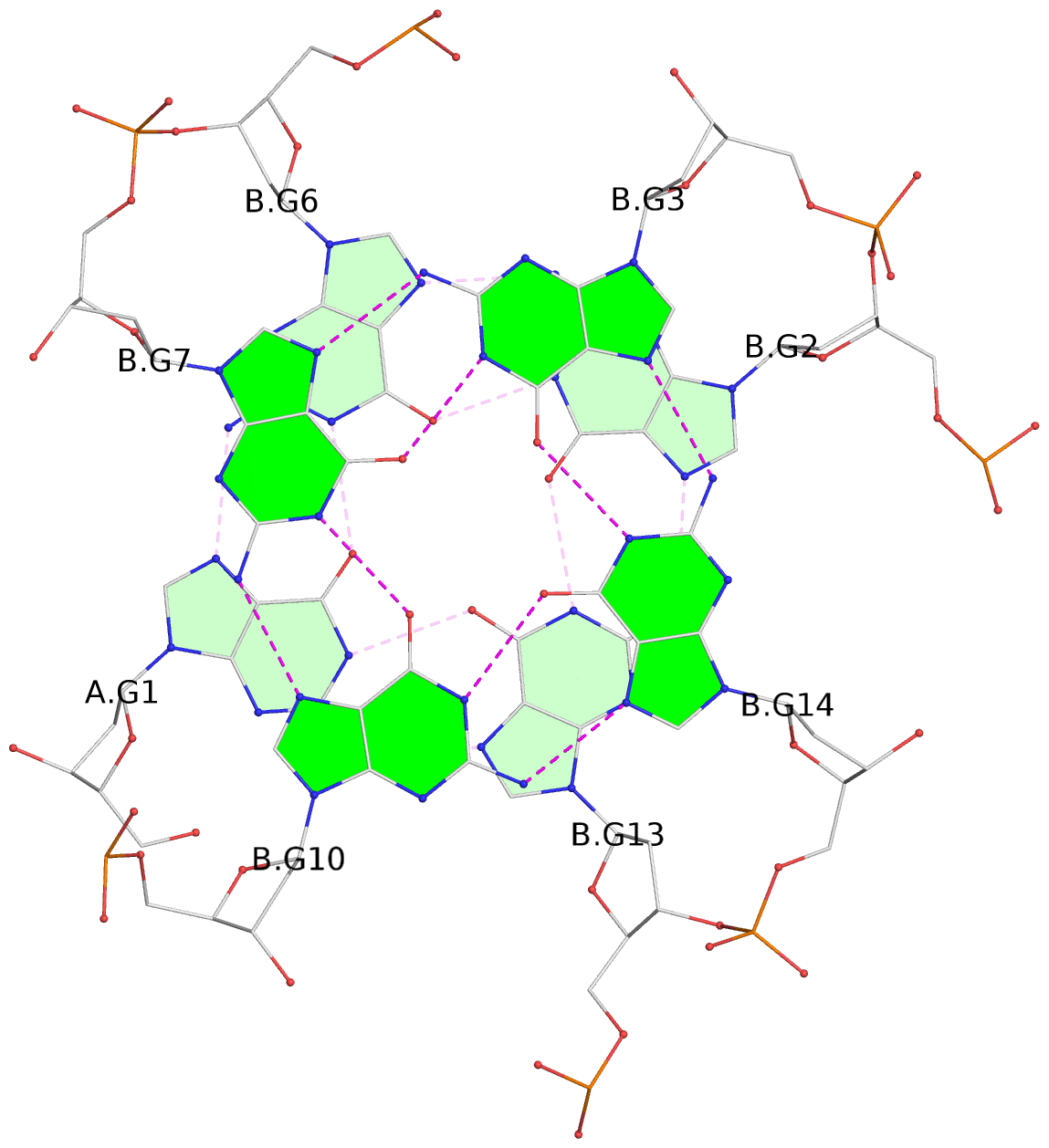
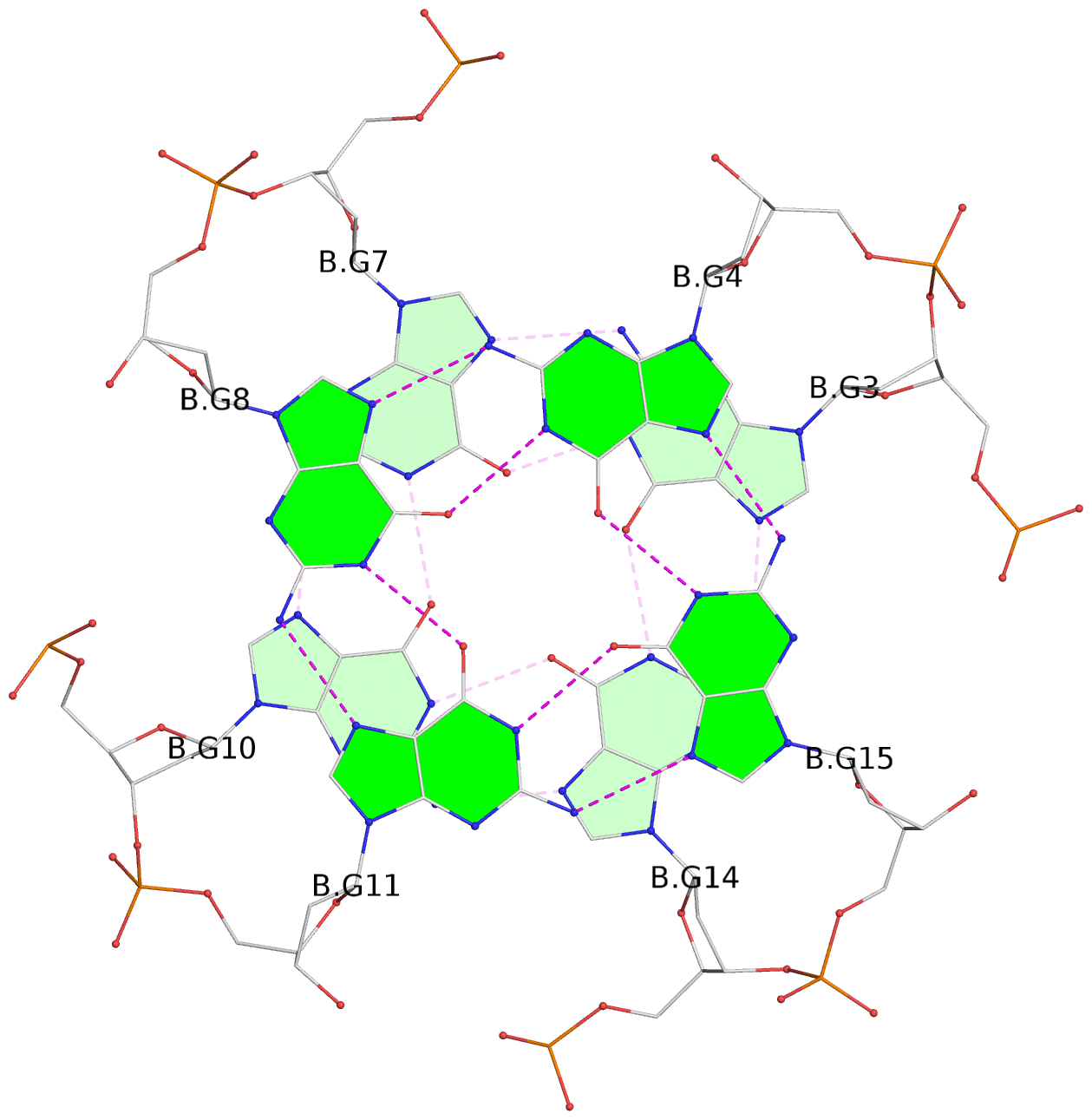
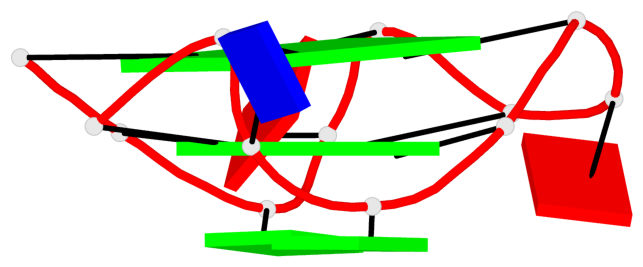
Base-block schematics in six views
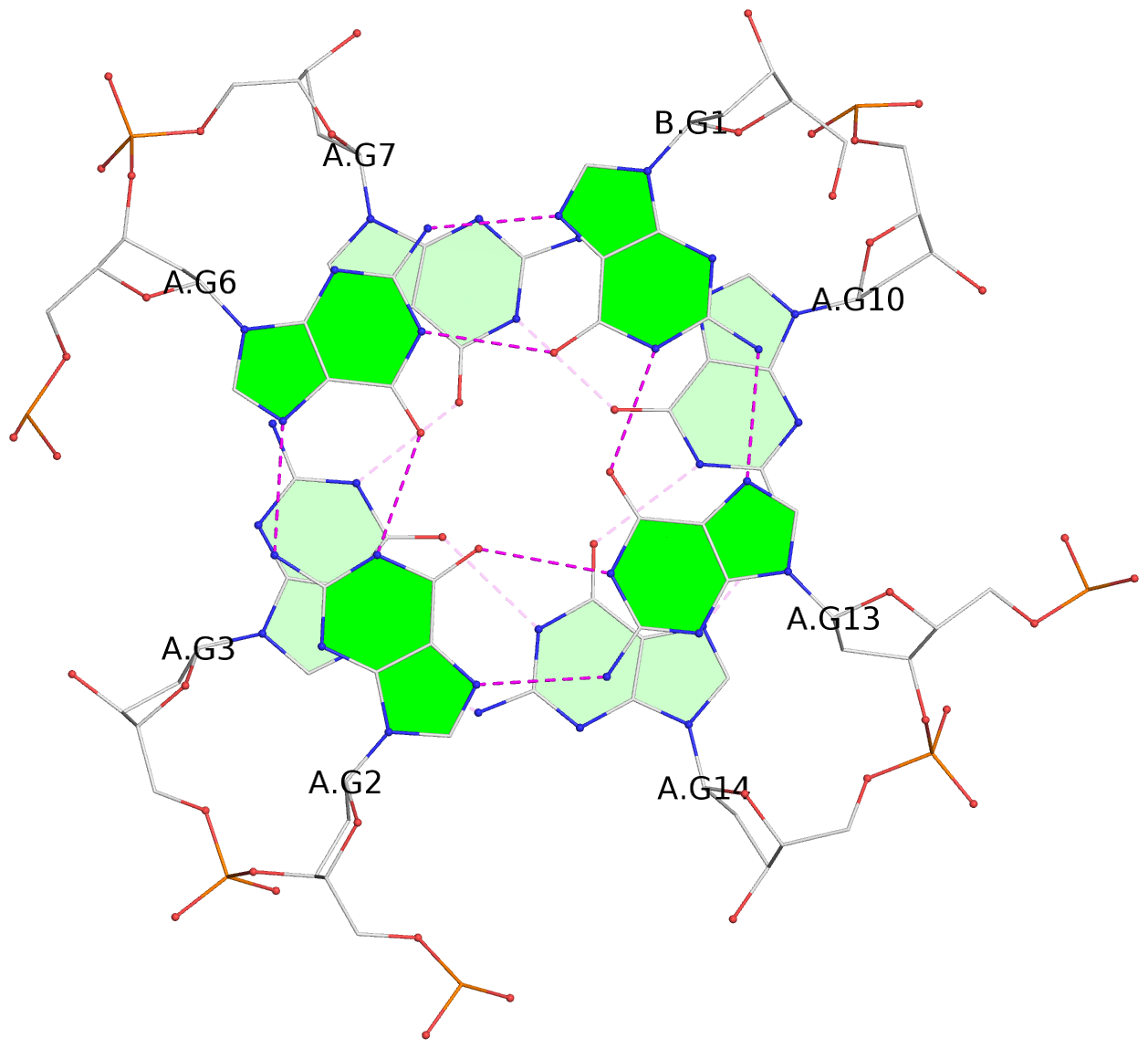
List of 6 G-tetrads
1 glyco-bond=s--- sugar=---. groove=w--n planarity=0.091 type=planar nts=4 GGGG A.DG1,B.DG6,B.DG2,B.DG13 2 glyco-bond=--s- sugar=---. groove=-wn- planarity=0.091 type=planar nts=4 GGGG A.DG2,A.DG6,B.DG1,A.DG13 3 glyco-bond=---- sugar=---- groove=---- planarity=0.125 type=planar nts=4 GGGG A.DG3,A.DG7,A.DG10,A.DG14 4 glyco-bond=---- sugar=---- groove=---- planarity=0.141 type=planar nts=4 GGGG A.DG4,A.DG8,A.DG11,A.DG15 5 glyco-bond=---- sugar=---- groove=---- planarity=0.126 type=planar nts=4 GGGG B.DG3,B.DG7,B.DG10,B.DG14 6 glyco-bond=---- sugar=---- groove=---- planarity=0.141 type=planar nts=4 GGGG B.DG4,B.DG8,B.DG11,B.DG15
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 6 G-tetrad layers, inter-molecular, with 2 stems
List of 2 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 2 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 2(-P-P-P), parallel(4+0)
Stem#2, 2 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 2(-P-P-P), parallel(4+0)
List of 1 G4 coaxial stack
1 G4 helix#1 contains 2 G4 stems: [#1,#2] [5'/5'-SEPARATED]