Detailed DSSR results for the G-quadruplex: PDB entry 230d
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 230d
- Class
- DNA
- Method
- NMR
- Summary
- Solution structures of unimolecular quadruplexes formed by oligonucleotides containing oxytricha telomere repeats
- Reference
- Smith FW, Schultze P, Feigon J (1995): "Solution structures of unimolecular quadruplexes formed by oligonucleotides containing Oxytricha telomere repeats." Structure, 3, 997-1008. doi: 10.1016/S0969-2126(01)00236-2.
- Abstract
- Background: Oligonucleotides containing the guanine-rich telomeric sequence of Oxytricha chromosomes (dT4G4) have previously been shown to form DNA quadruplexes comprising guanine quartets stabilized by cations. Two different structures have been reported for both d(G4T4G4) (Oxy1.5) and d(G4T4G4T4G4T4G4) (Oxy3.5).
Results: Here we present the solution structure of a uracil- and inosine-containing derivative of Oxy3.5, d(G4TUTUG4T4G4UUTTG3I) (Oxy3.5-U4128), determined using two-dimensional 1H and 31P NMR techniques. This oligonucleotide forms a unimolecular quadruplex that is very similar to the dimeric Oxy1.5 solution structure, in that it contains a loop spanning the diagonal of an end quartet. The groove widths, strand polarities, and positions of the syn bases along the G4 tracts and within the quartets are all as reported for Oxy1.5. The first and third pyrimidine tracts form parallel loops spanning a wide groove and a narrow groove respectively.
Conclusions: Both Oxy3.5 and Oxy3.5-U(4)128 form unimolecular quadruplexes in solution with a diagonal central T4 loop. These results conflict with those reported for d(G4TUTUG4TTUUG4UUTTG4) in solution, in which the central loop spans a wide groove. - G4 notes
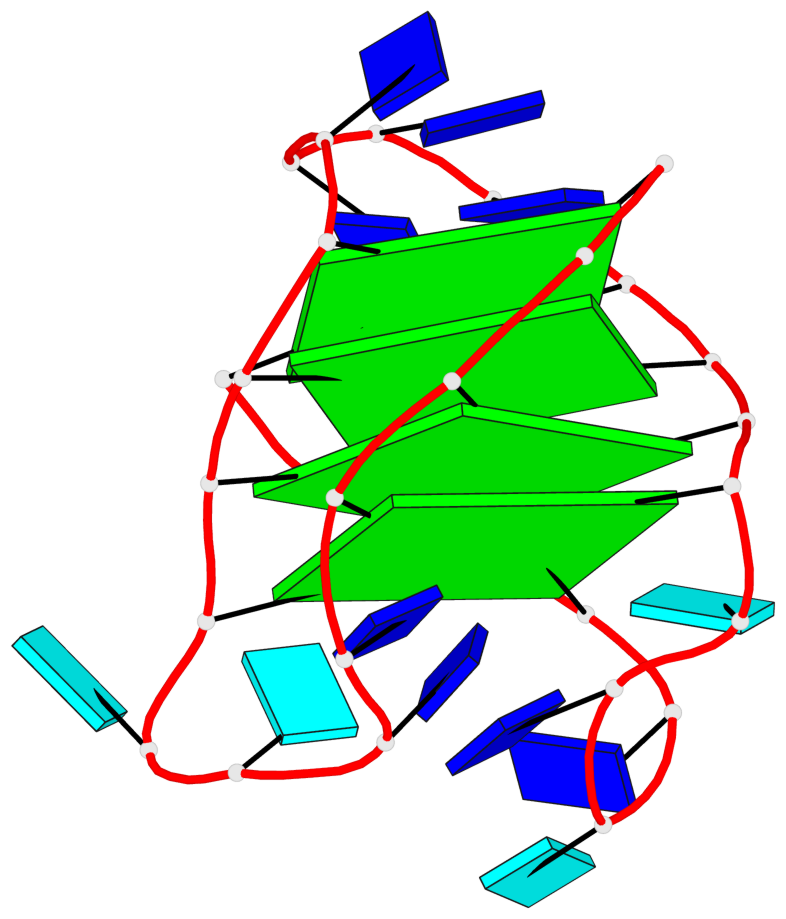
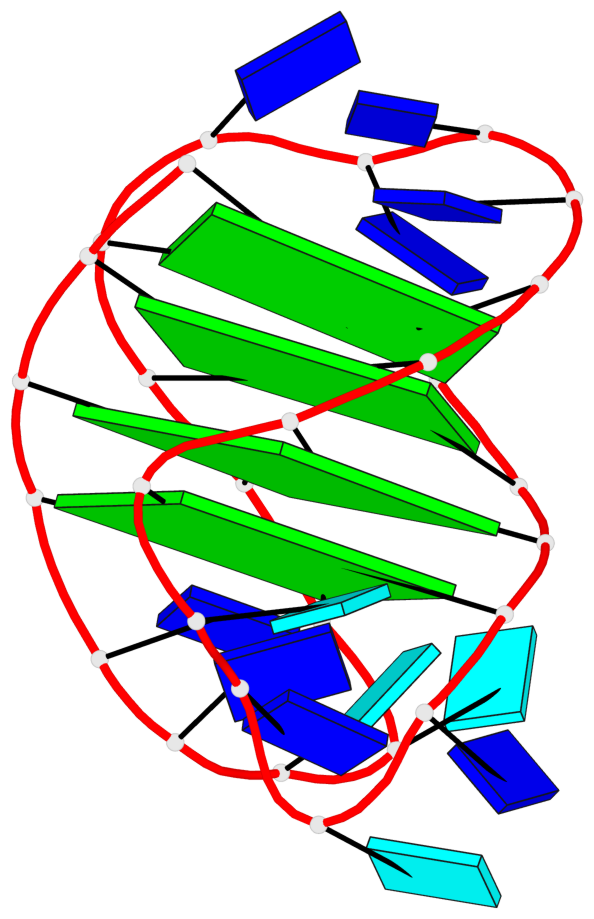
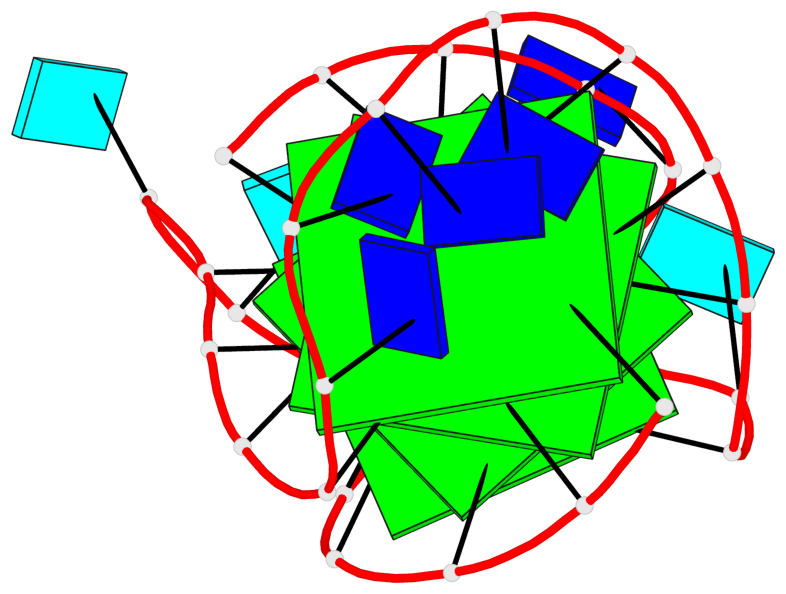
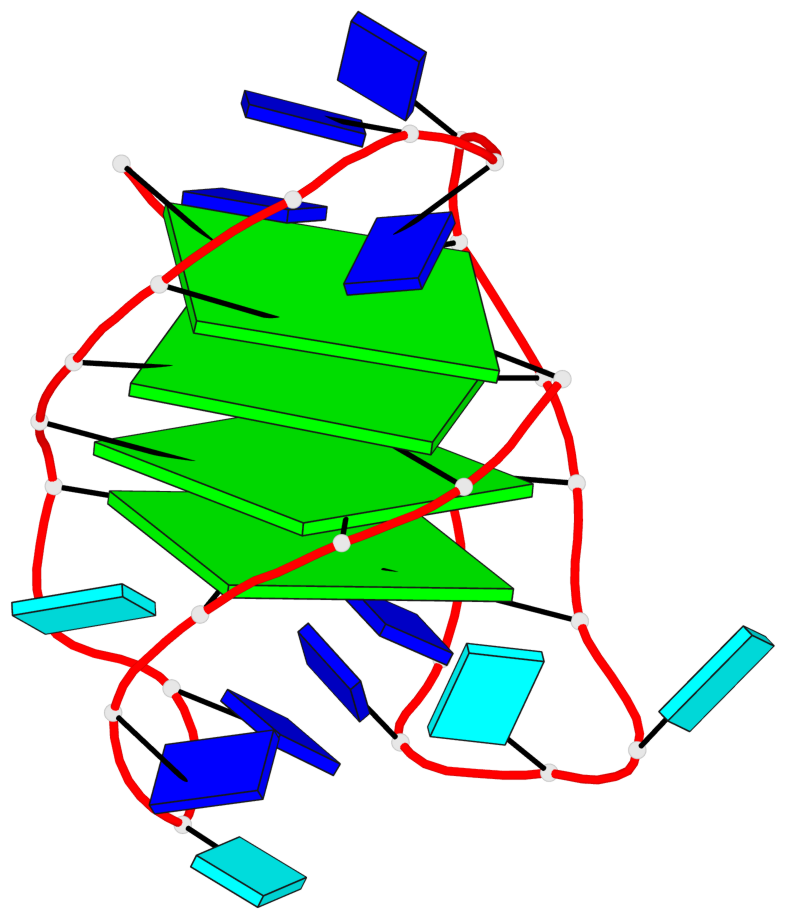
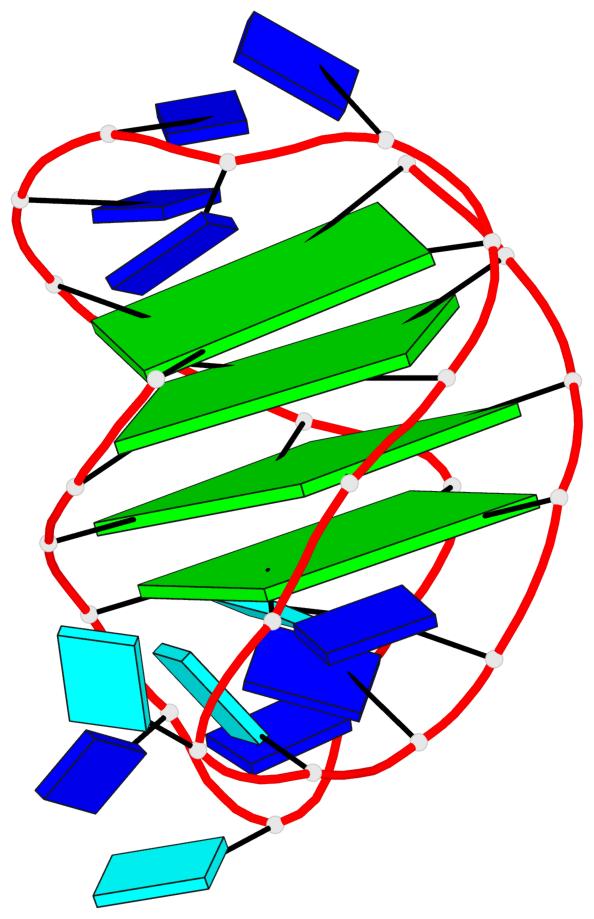
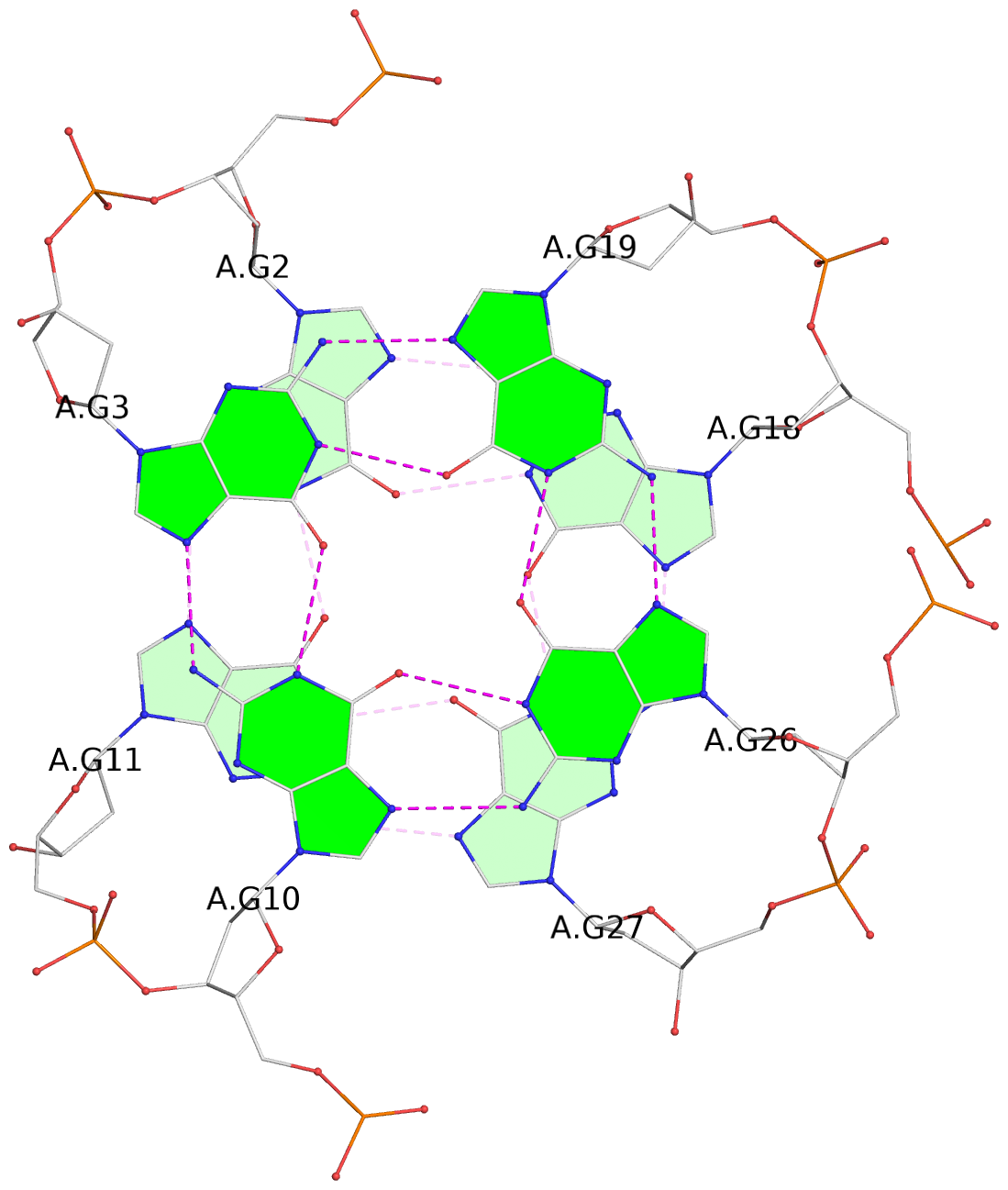
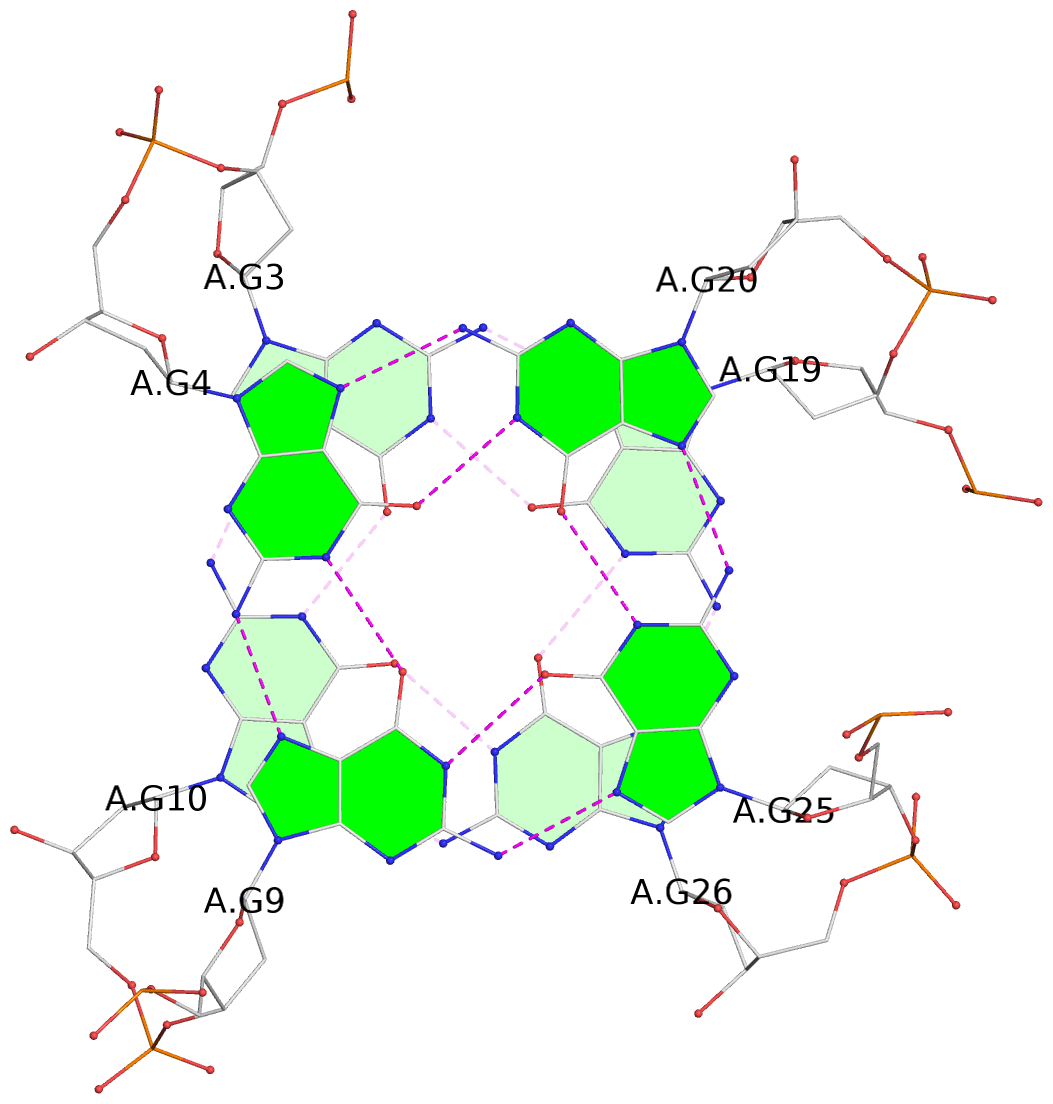
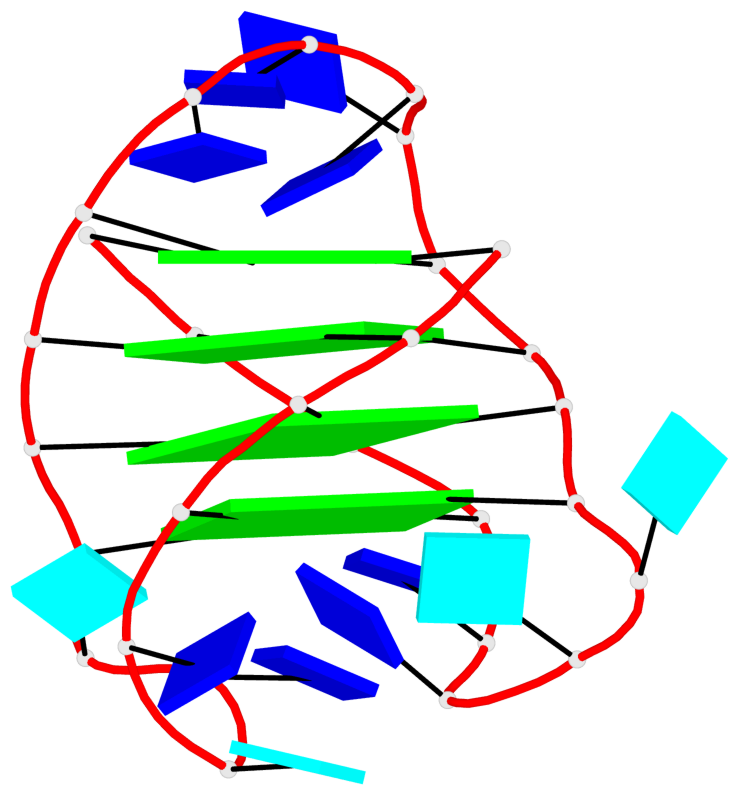
- 4 G-tetrads, 1 G4 helix, 1 G4 stem, 4(-LwD+Ln), basket(2+2), UDDU
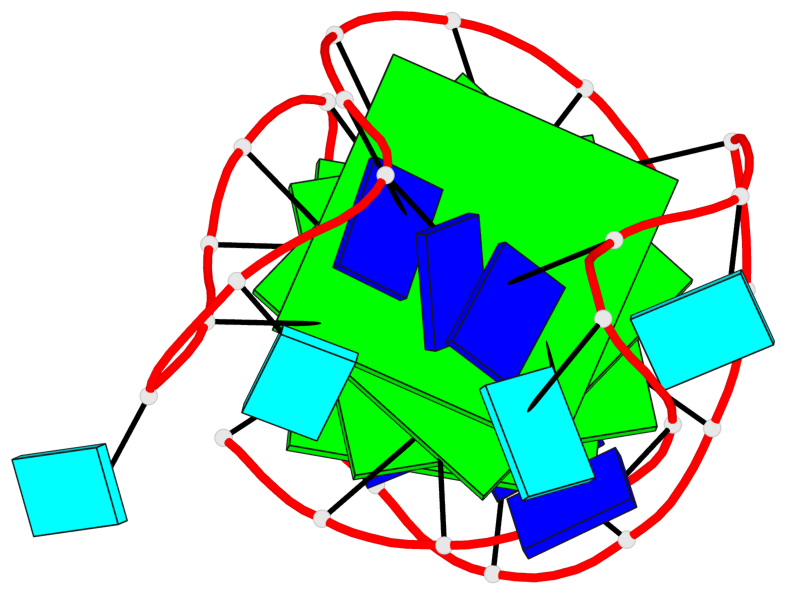
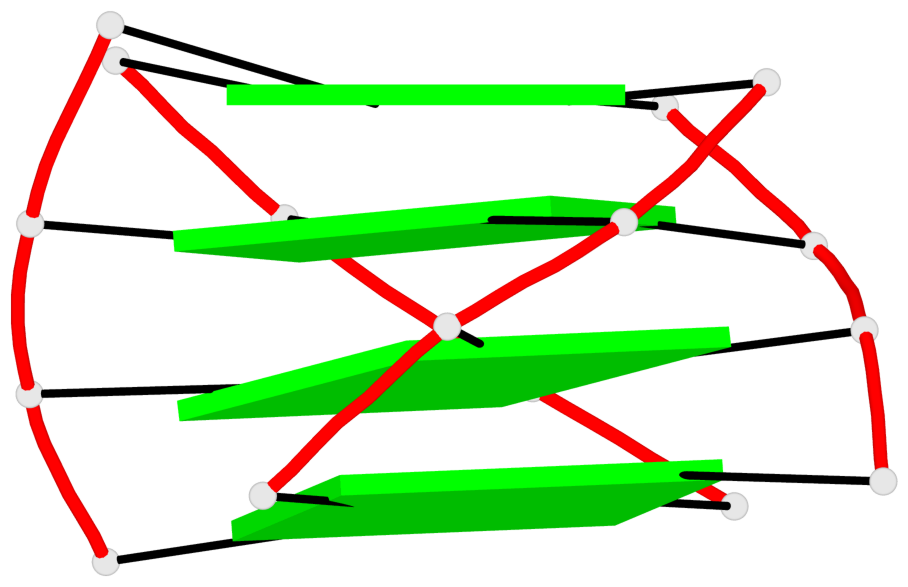
Base-block schematics in six views
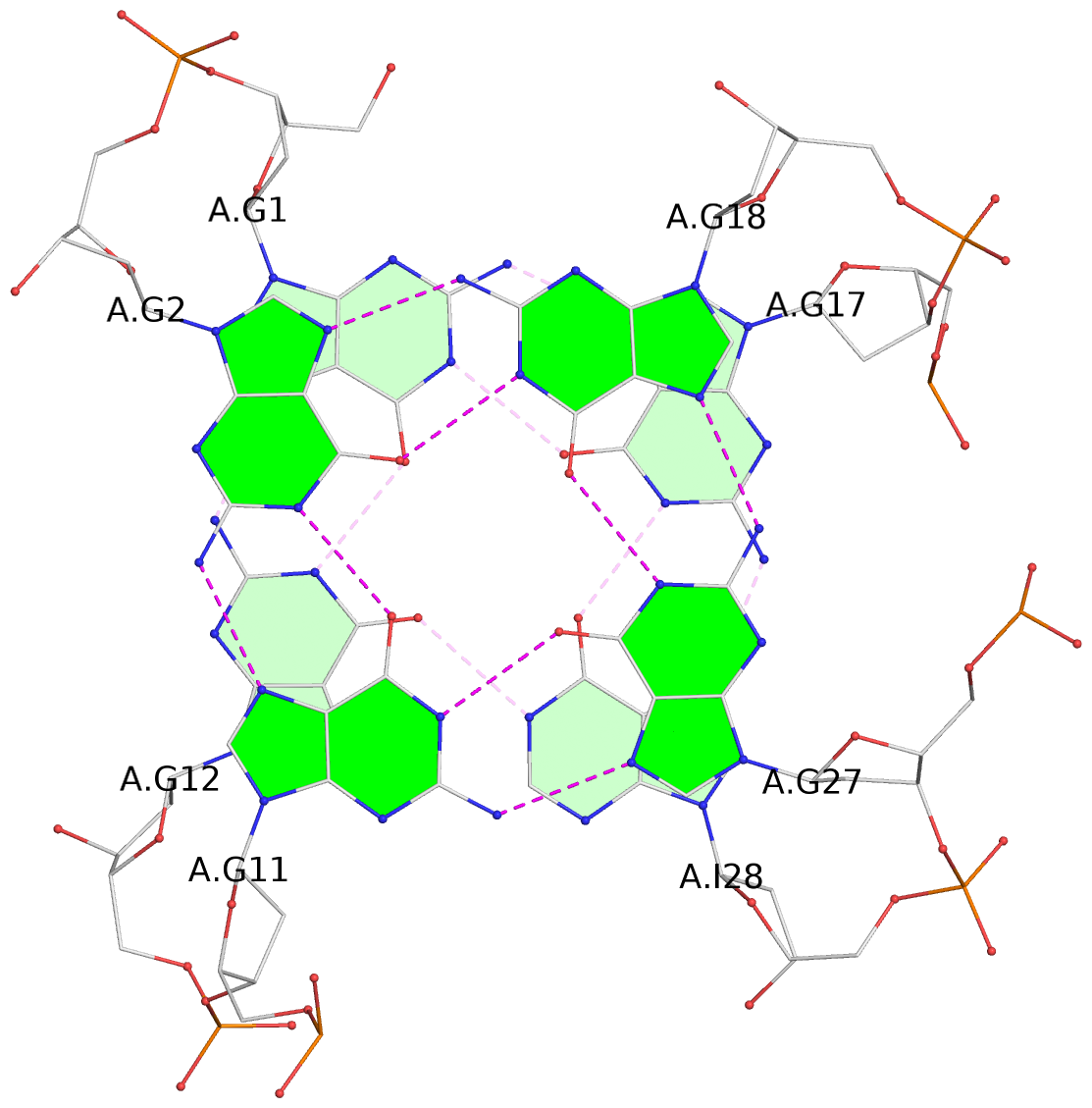
List of 4 G-tetrads
1 glyco-bond=s--s sugar=---- groove=w-n- planarity=0.109 type=planar nts=4 GGgG A.DG1,A.DG12,A.DI28,A.DG17 2 glyco-bond=-ss- sugar=---- groove=w-n- planarity=0.047 type=planar nts=4 GGGG A.DG2,A.DG11,A.DG27,A.DG18 3 glyco-bond=s--s sugar=---- groove=w-n- planarity=0.084 type=planar nts=4 GGGG A.DG3,A.DG10,A.DG26,A.DG19 4 glyco-bond=-ss- sugar=---. groove=w-n- planarity=0.088 type=planar nts=4 GGGG A.DG4,A.DG9,A.DG25,A.DG20
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 4 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.