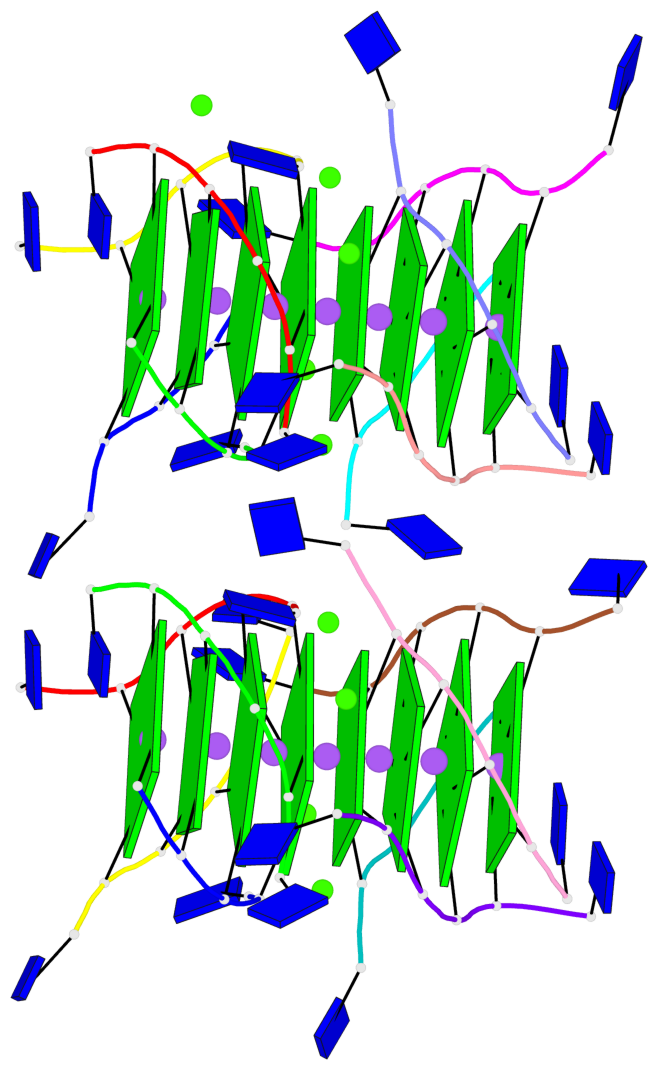
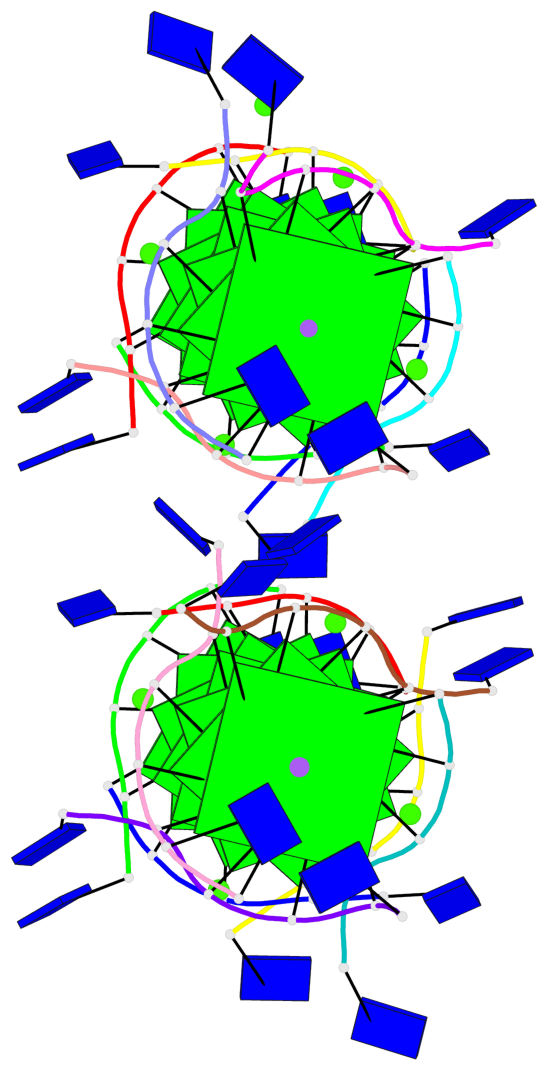
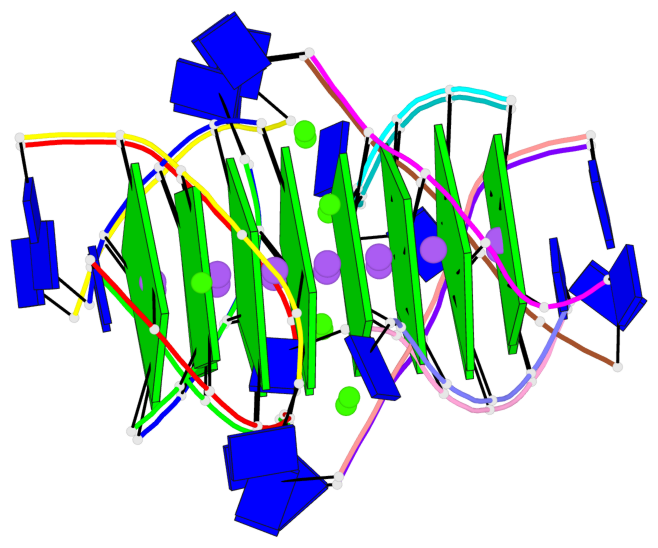
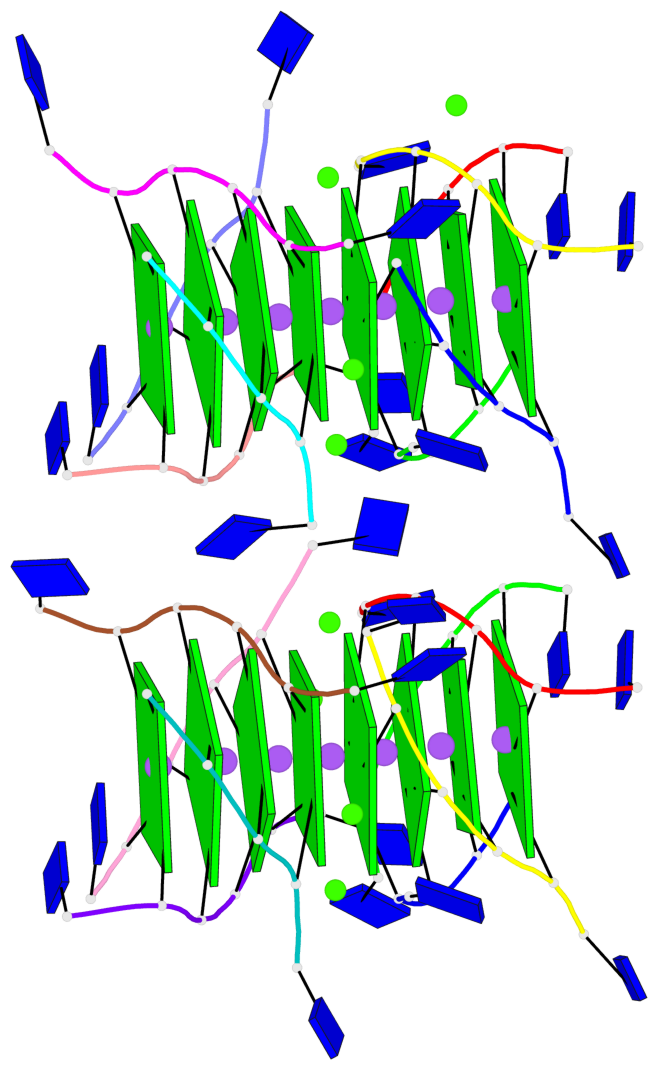
Detailed DSSR results for the G-quadruplex: PDB entry 244d
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 244d
- Class
- DNA
- Method
- X-ray (1.2 Å)
- Summary
- The high-resolution crystal structure of a parallel-stranded guanine tetraplex
- Reference
- Laughlan G, Murchie AI, Norman DG, Moore MH, Moody PC, Lilley DM, Luisi B (1994): "The high-resolution crystal structure of a parallel-stranded guanine tetraplex." Science, 265, 520-524.
- Abstract
- Repeat tracts of guanine bases found in DNA and RNA can form tetraplex structures in the presence of a variety of monovalent cations. Evidence suggests that guanine tetraplexes assume important functions within chromosomal telomeres, immunoglobulin switch regions, and the human immunodeficiency virus genome. The structure of a parallel-stranded tetraplex formed by the hexanucleotide d(TG4T) and stabilized by sodium cations was determined by x-ray crystallography to 1.2 angstroms resolution. Sharply resolved sodium cations were found between and within planes of hydrogen-bonded guanine quartets, and an ordered groove hydration was observed. Distinct intra- and intermolecular stacking arrangements were adopted by the guanine quartets. Thymine bases were exclusively involved in making extensive lattice contacts.
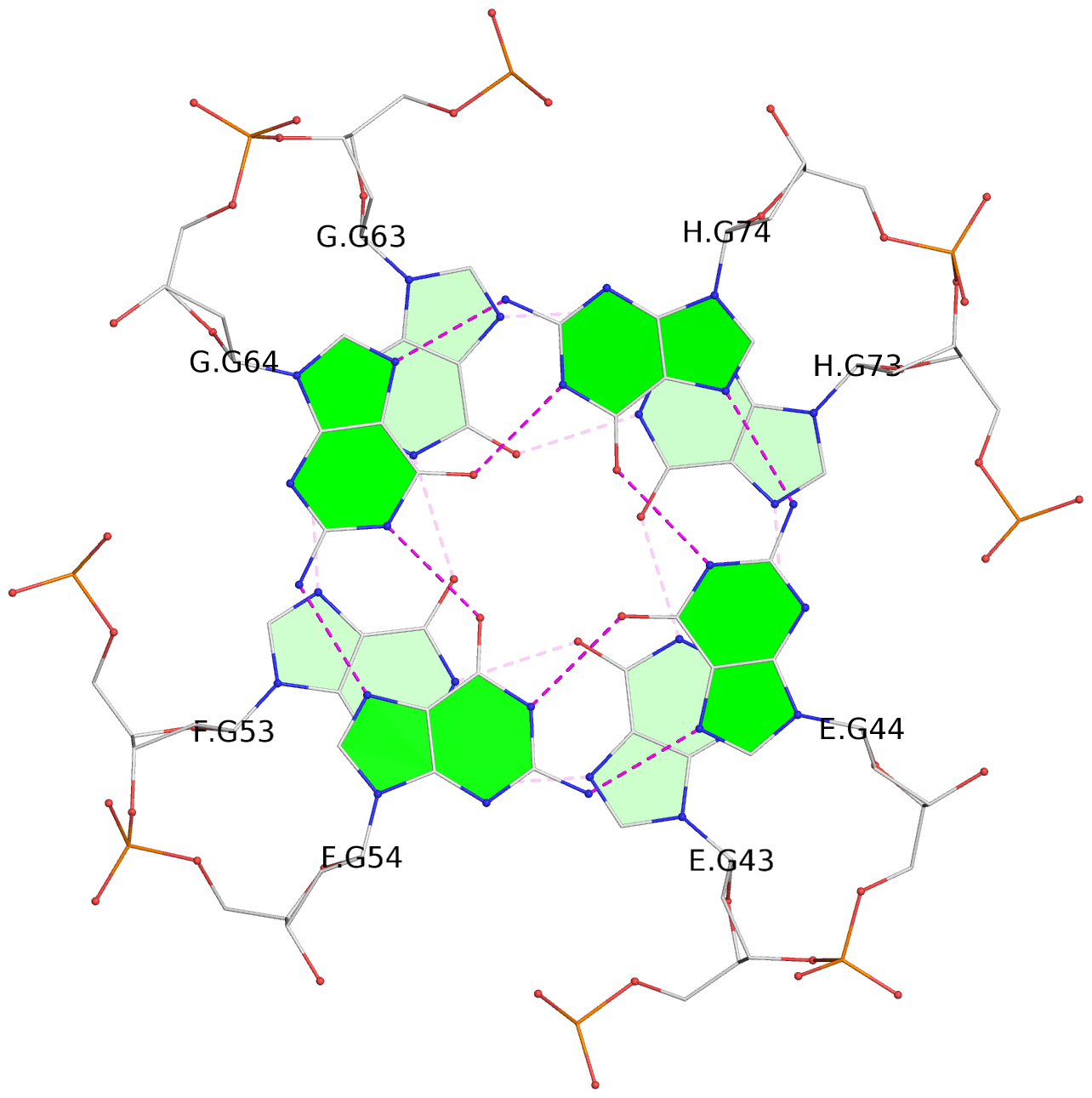
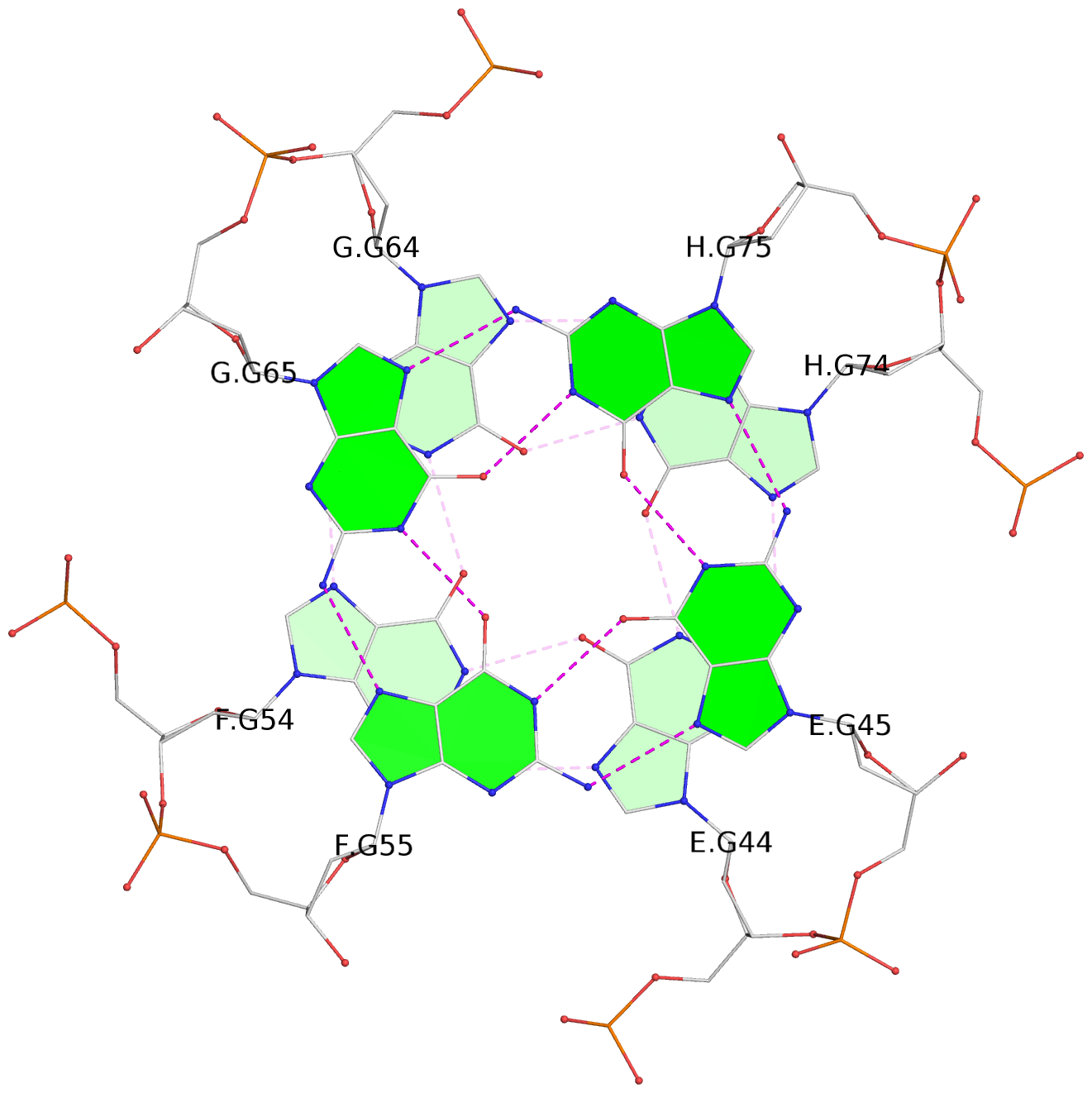
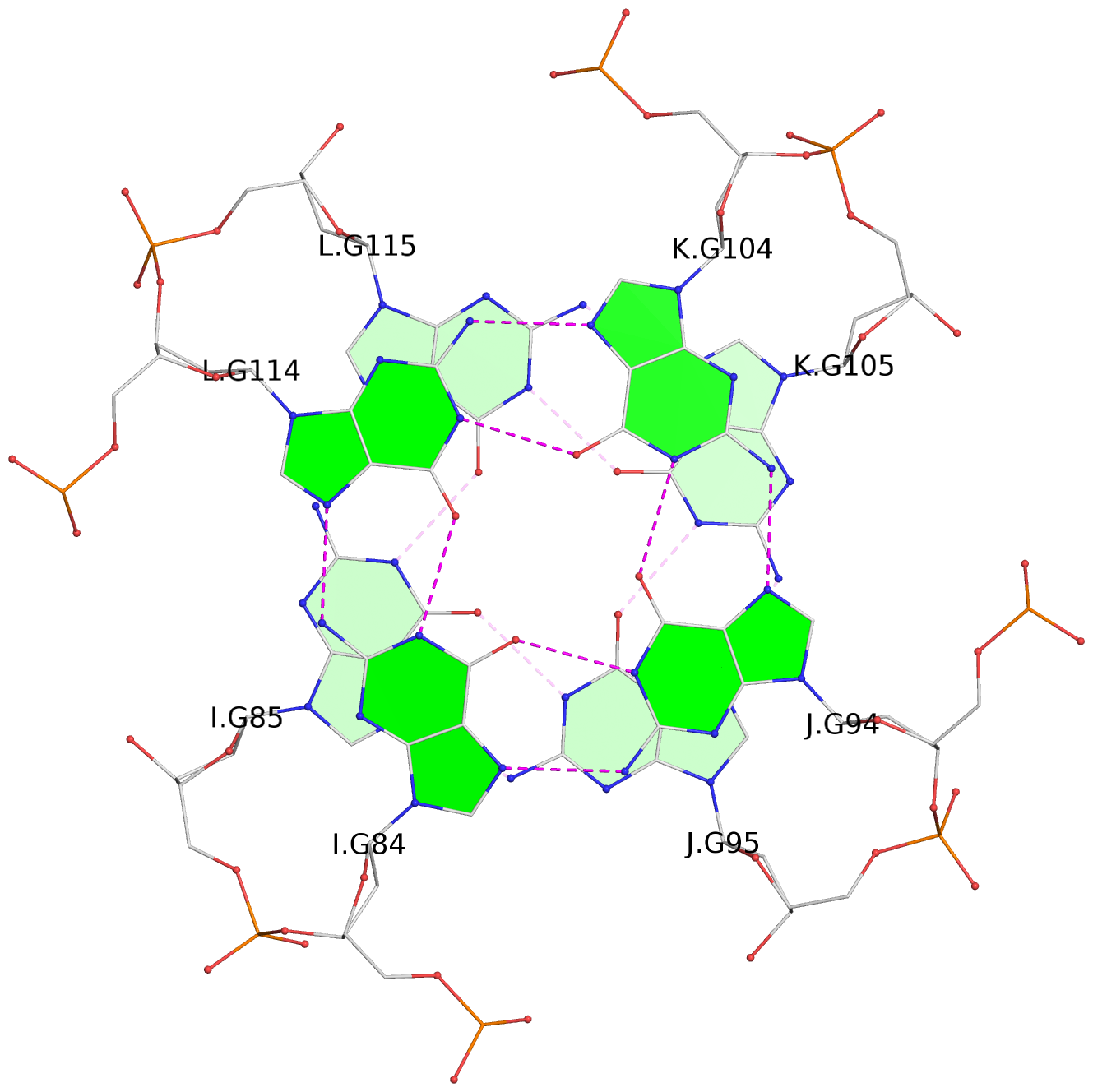
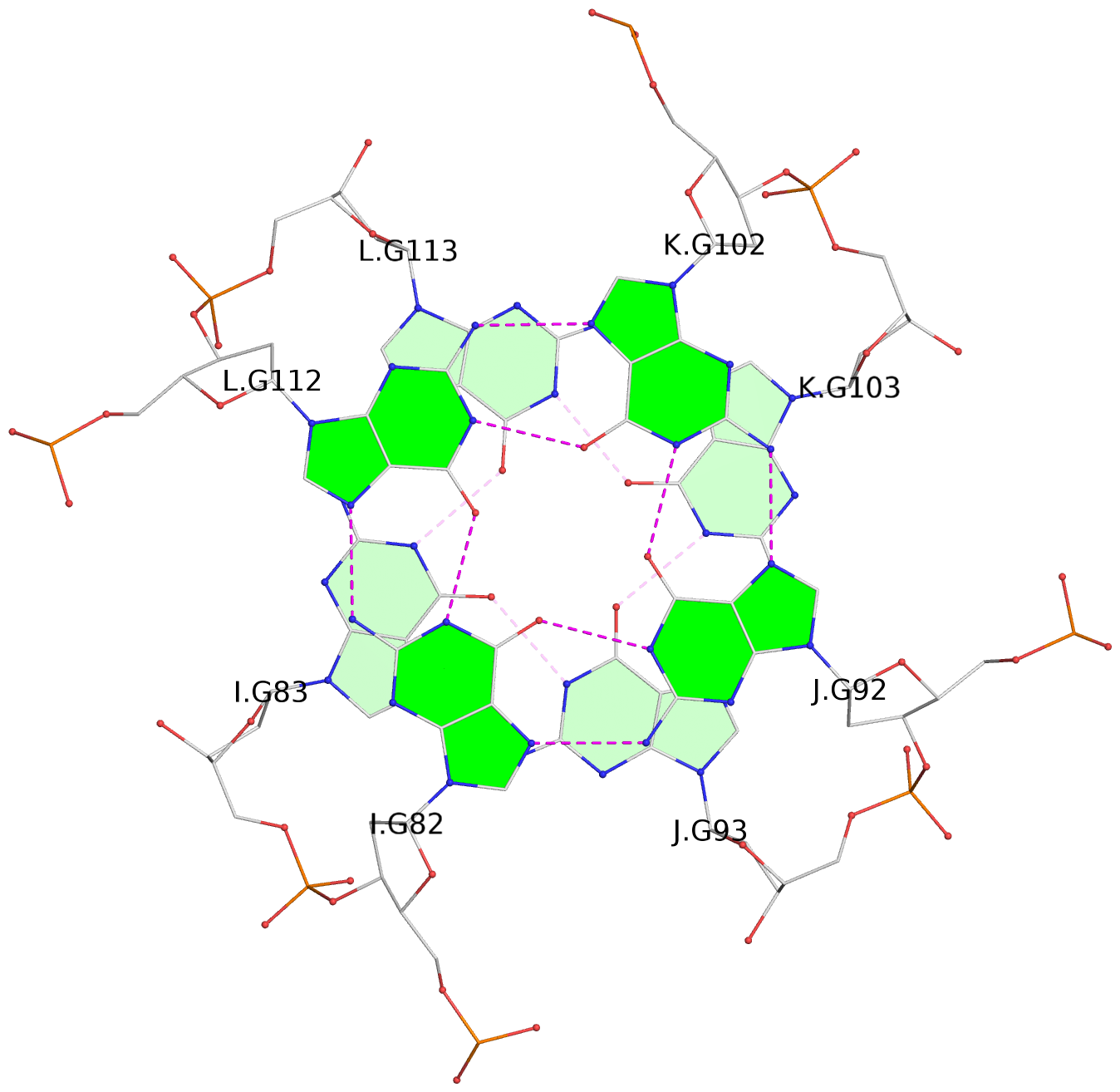
- G4 notes
- 16 G-tetrads, 2 G4 helices, 4 G4 stems, 2 G4 coaxial stacks, parallel(4+0), UUUU, coaxial interfaces: 5'/5'
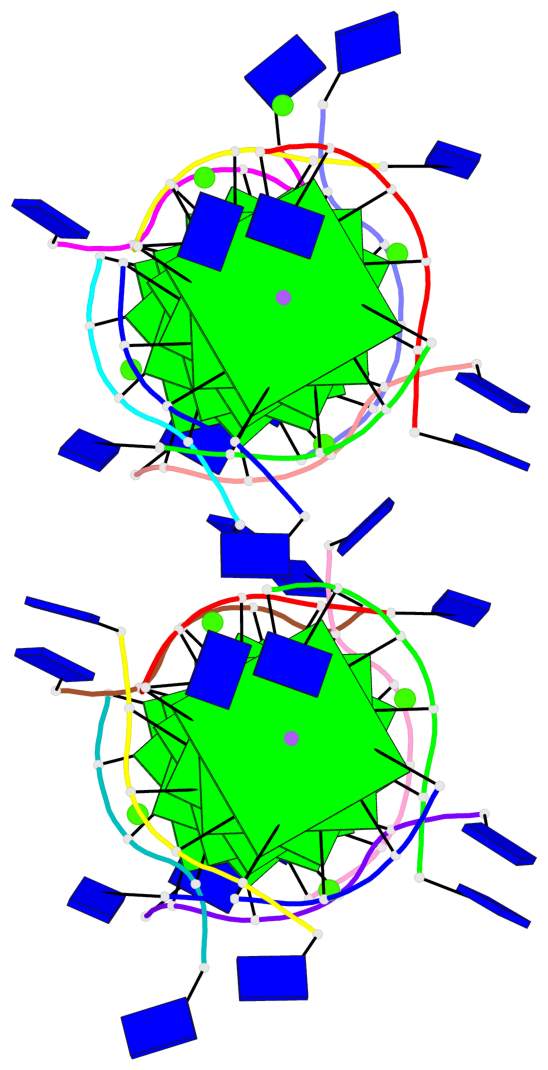
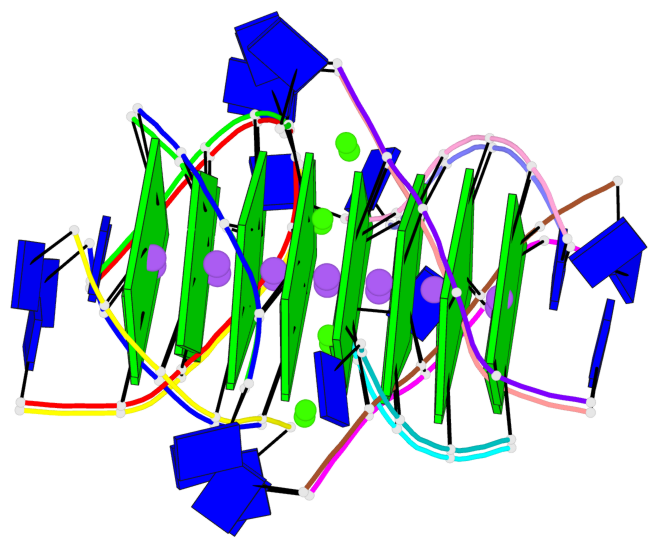
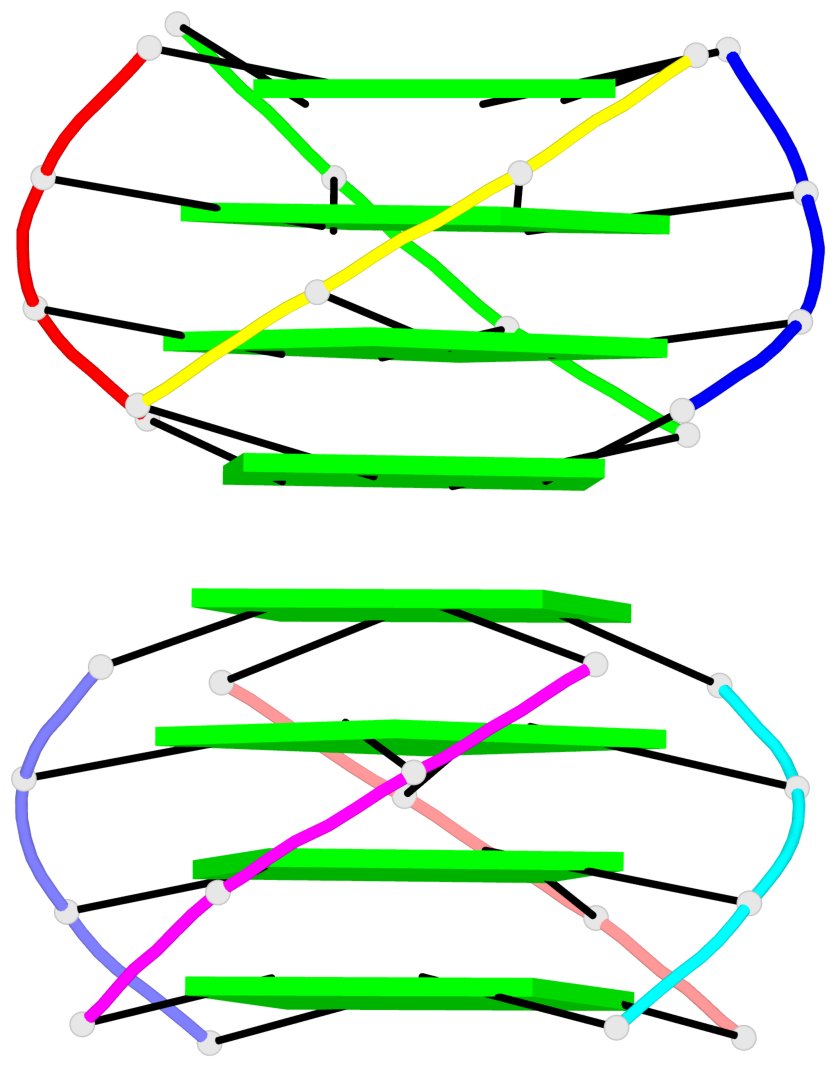
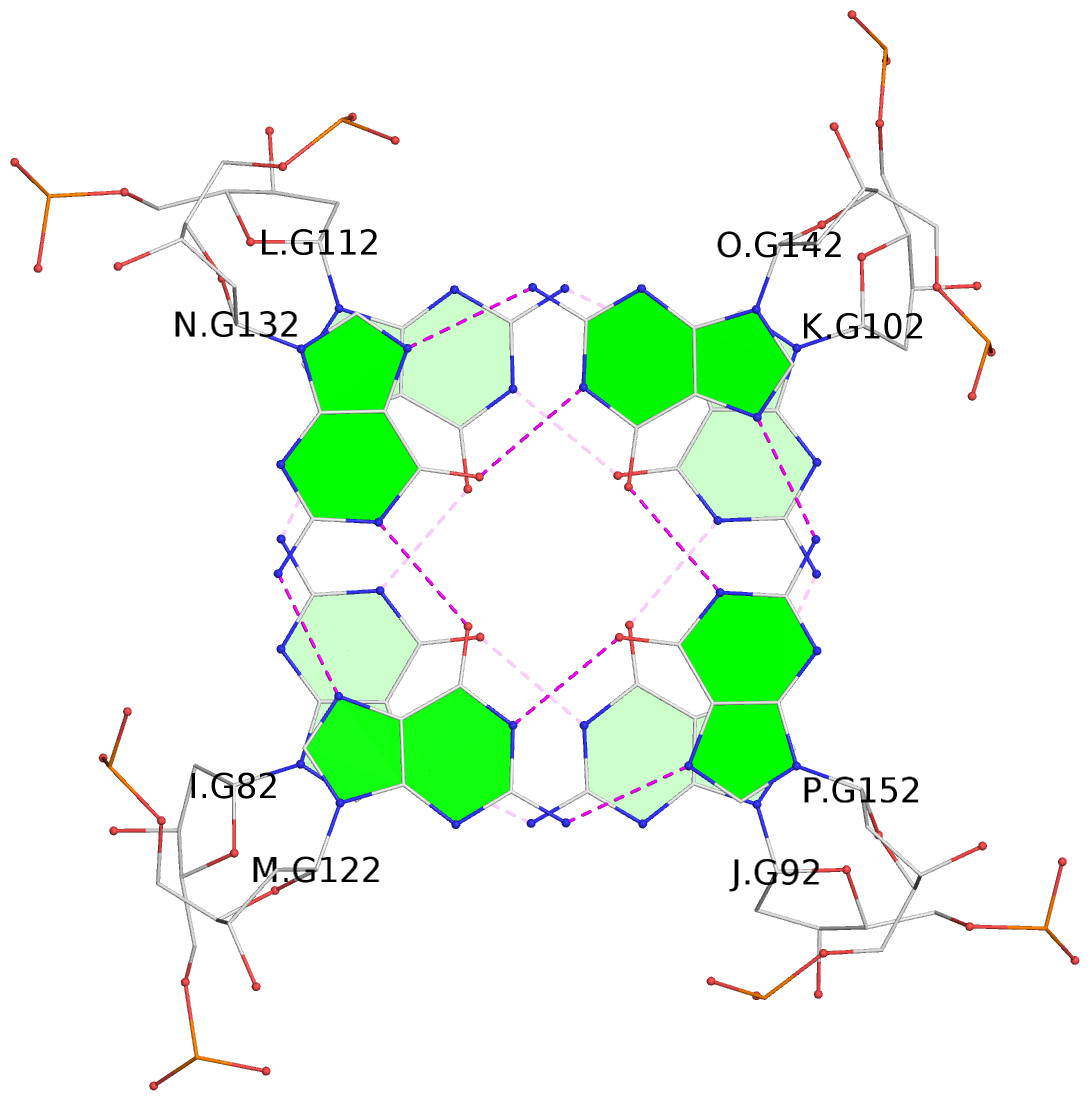
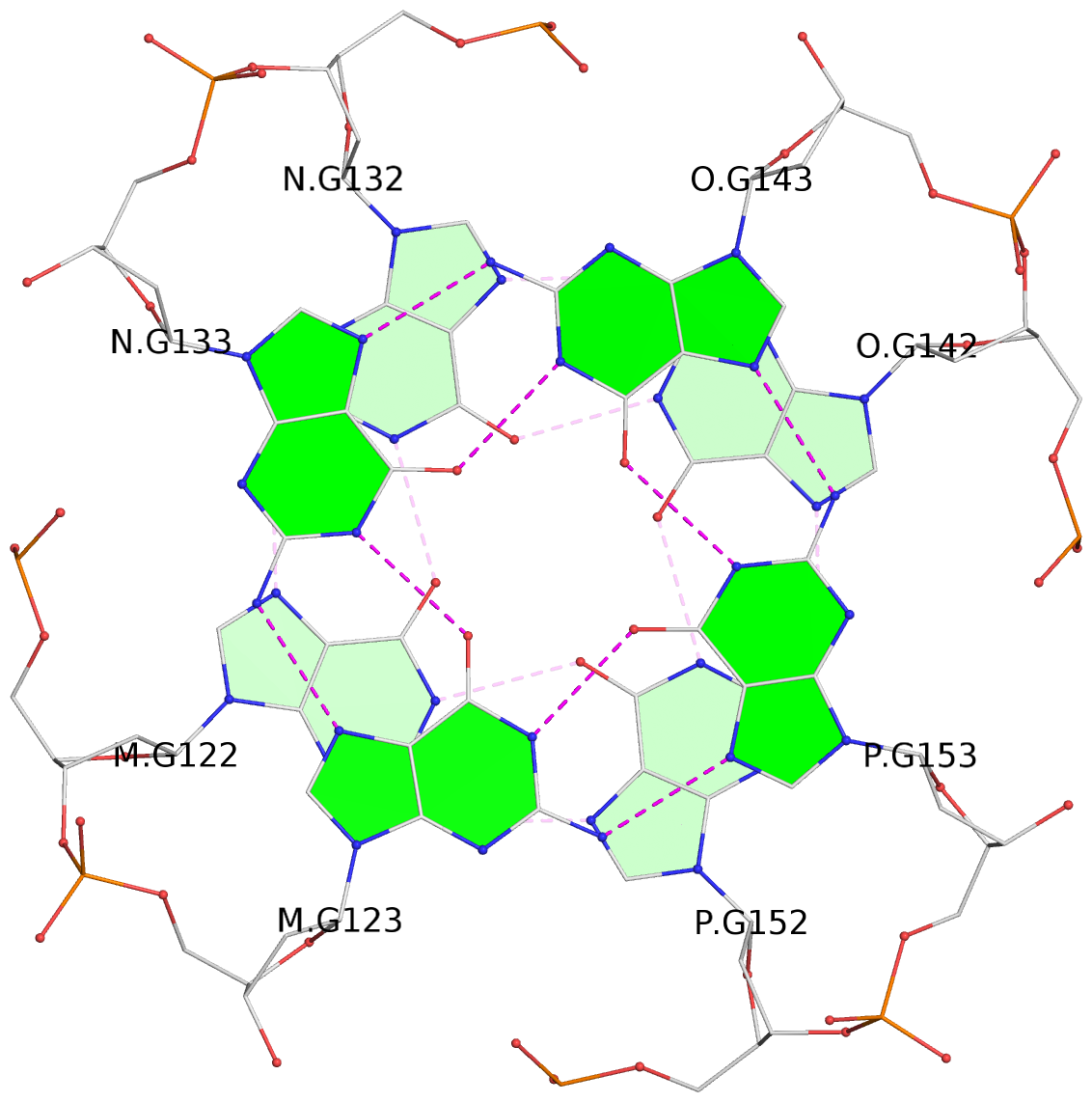
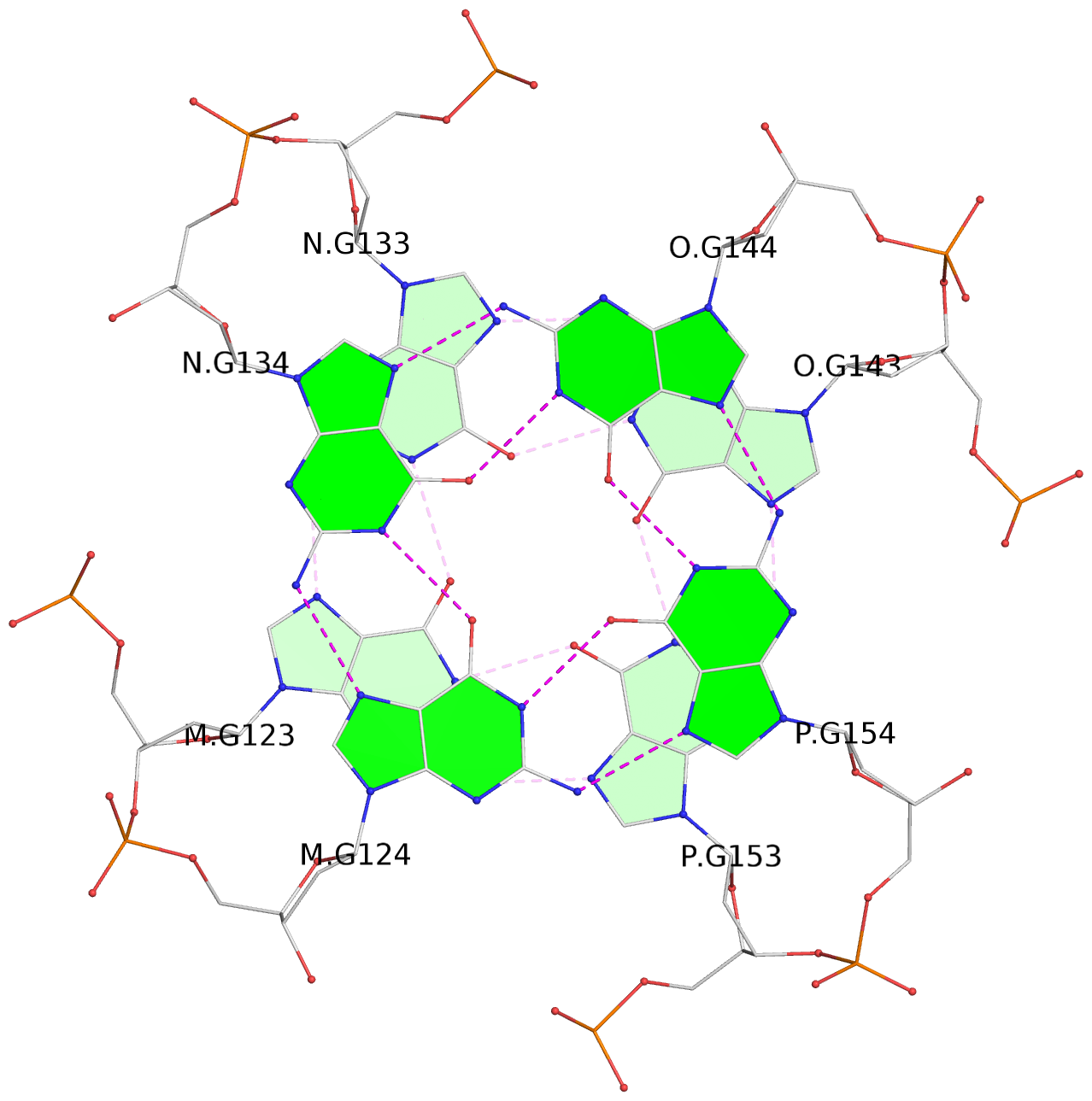
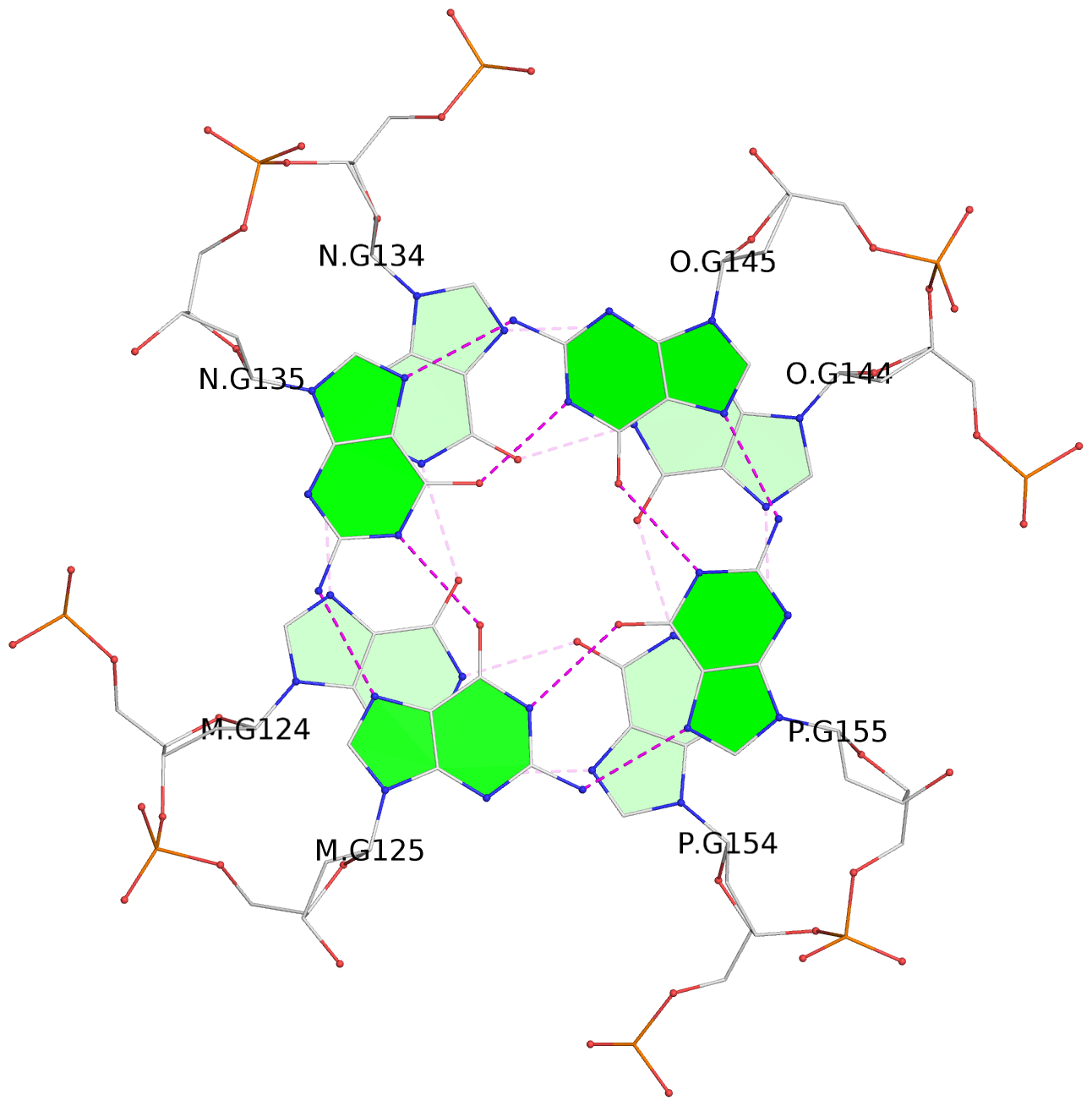
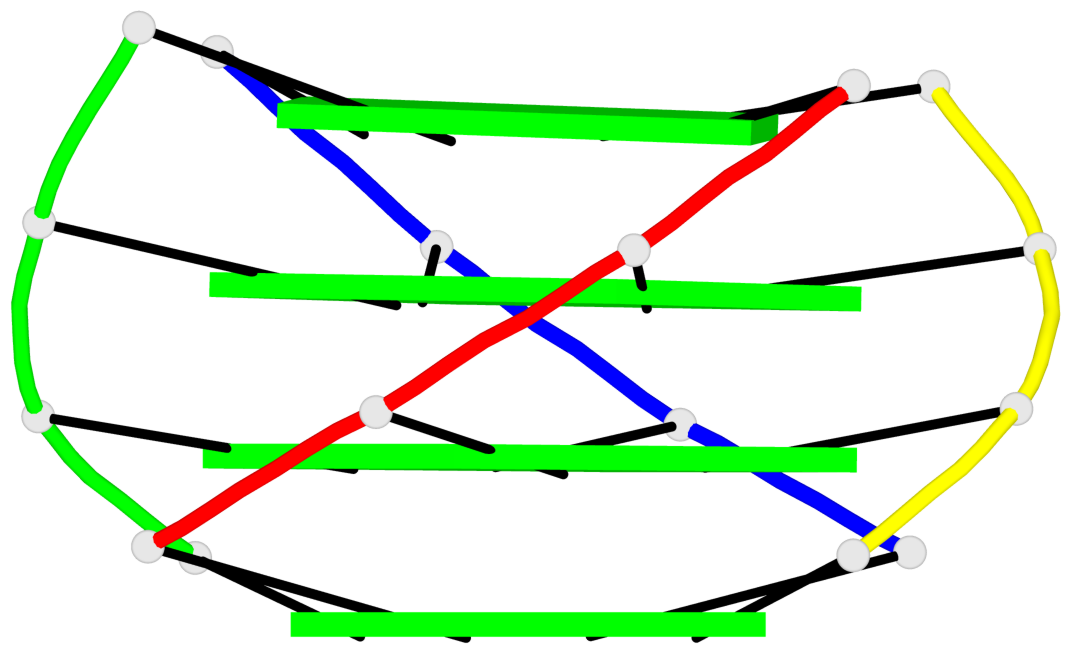
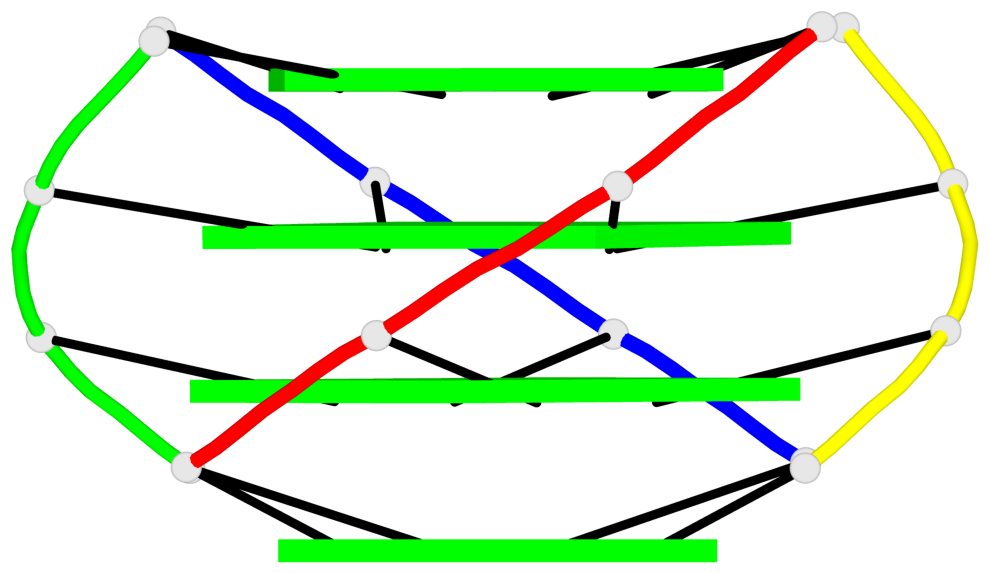
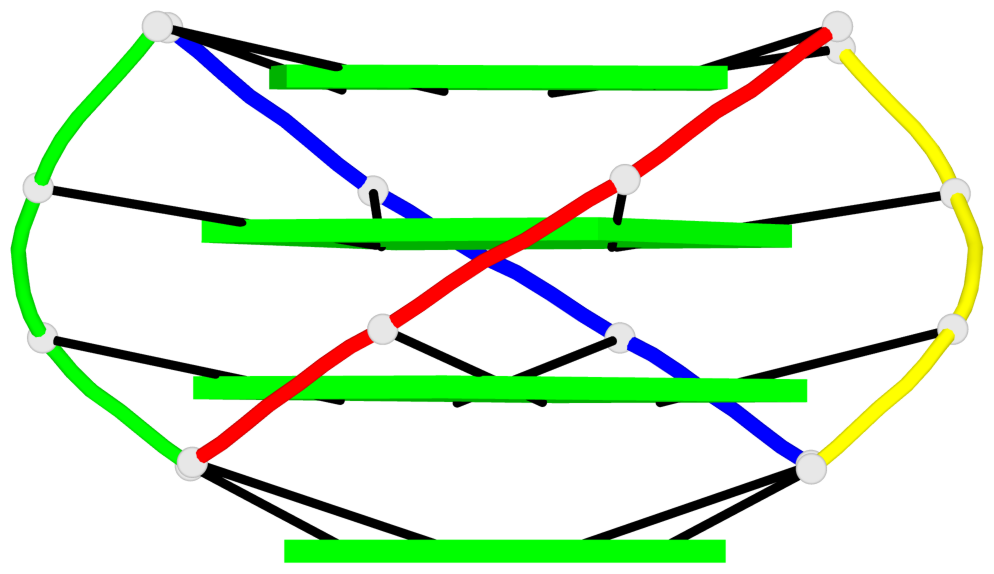
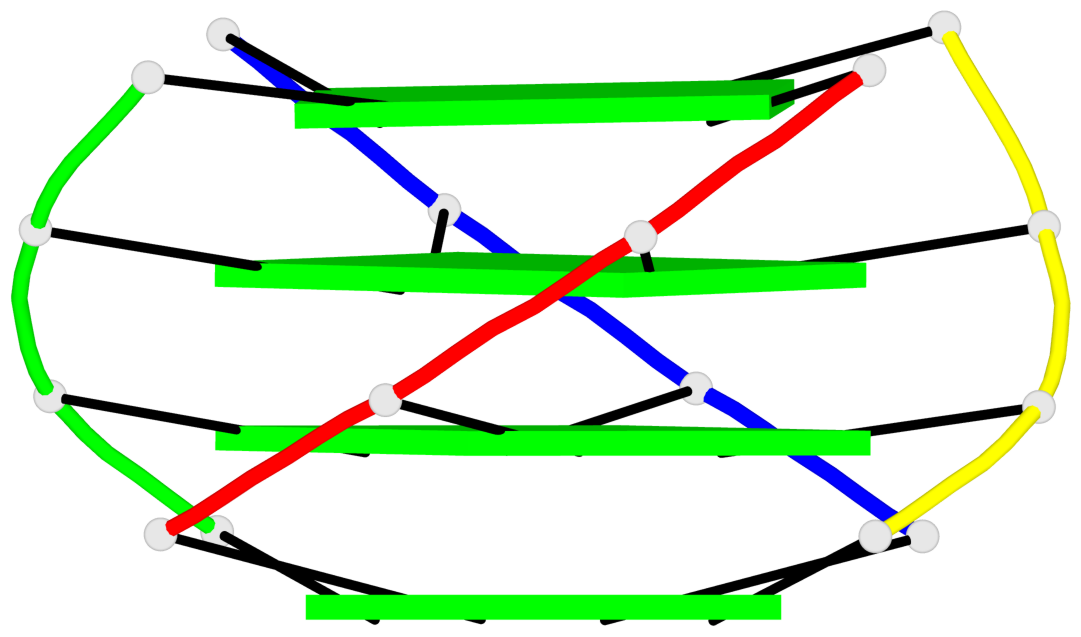
Base-block schematics in six views
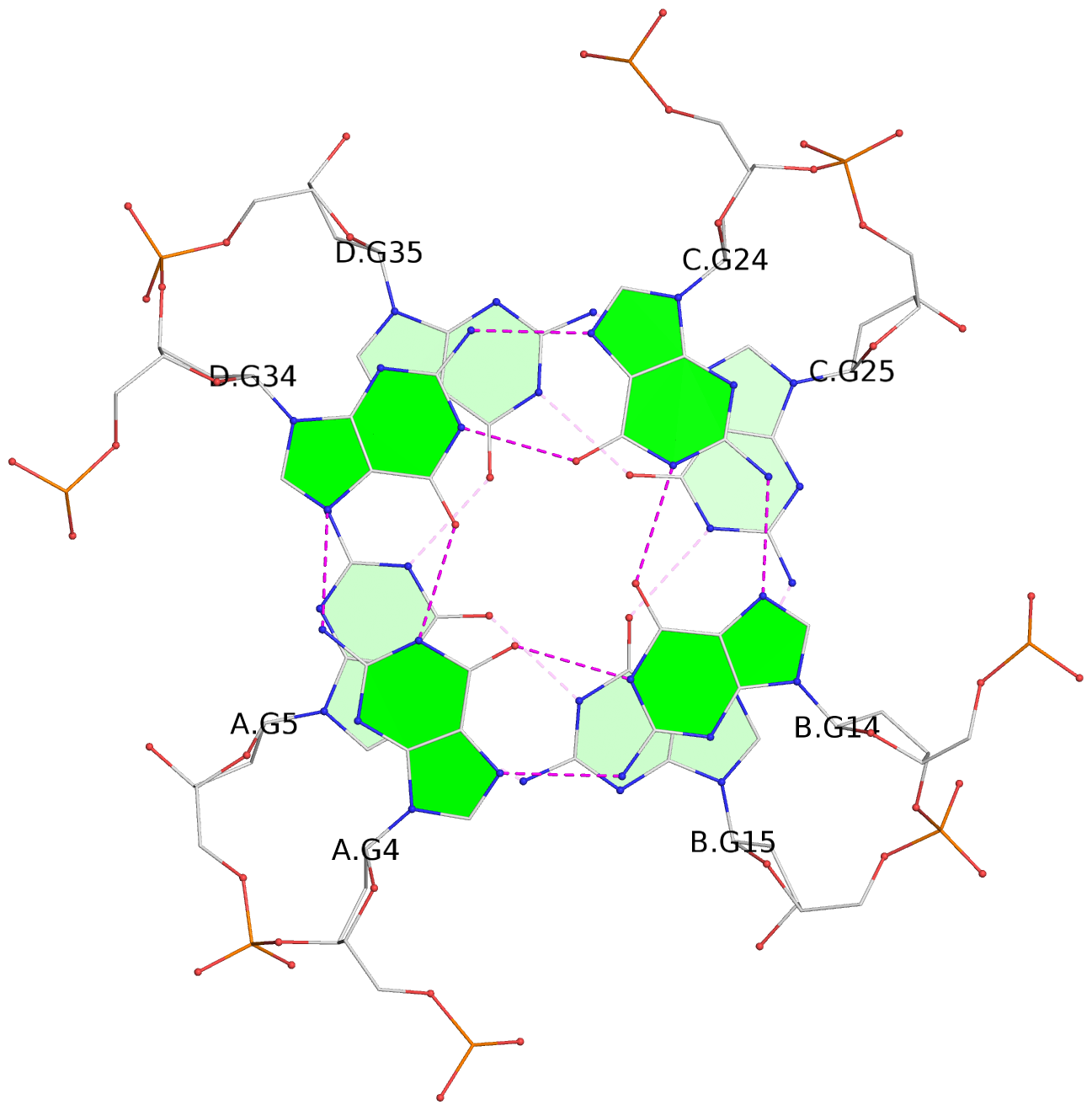
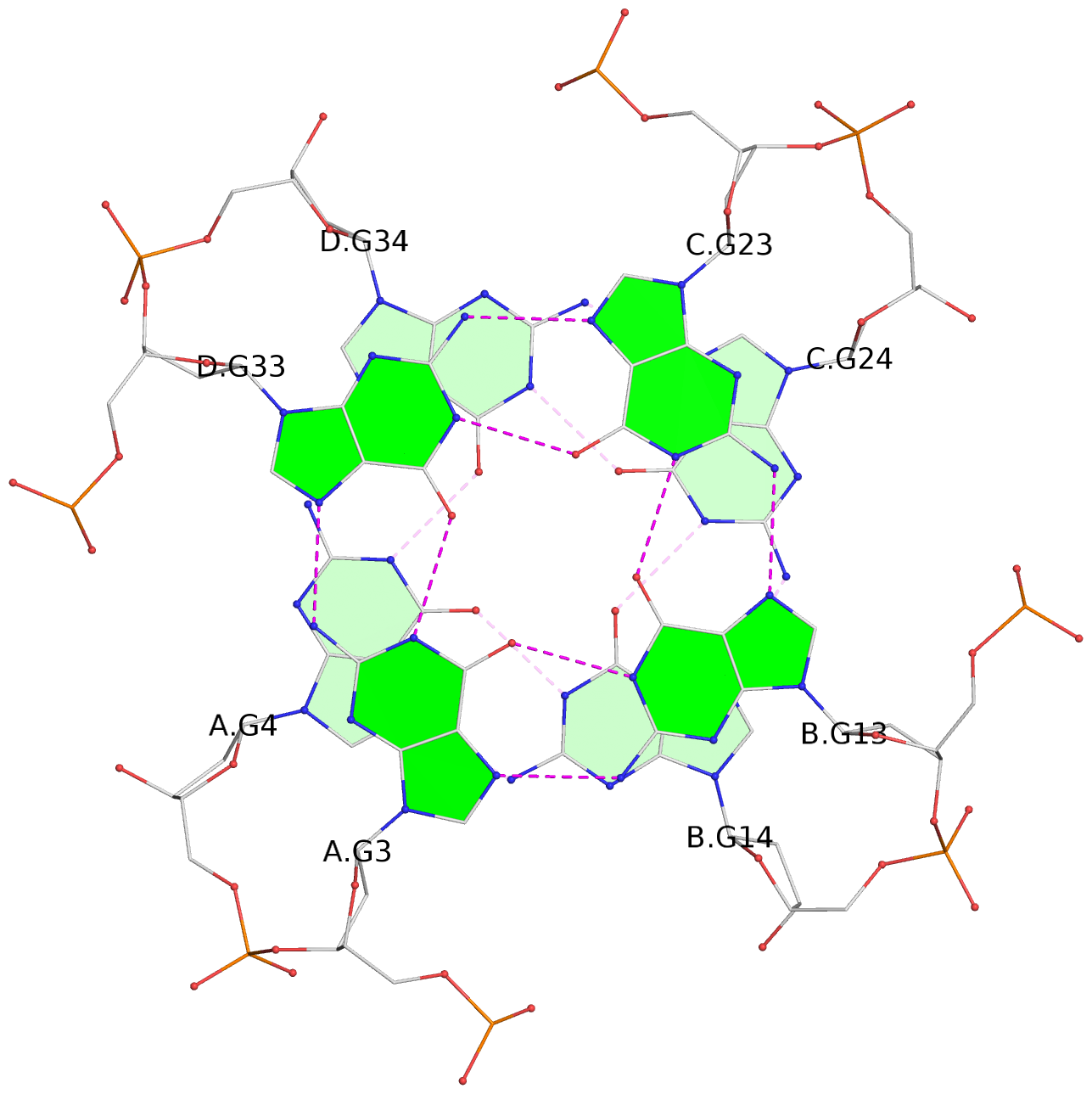
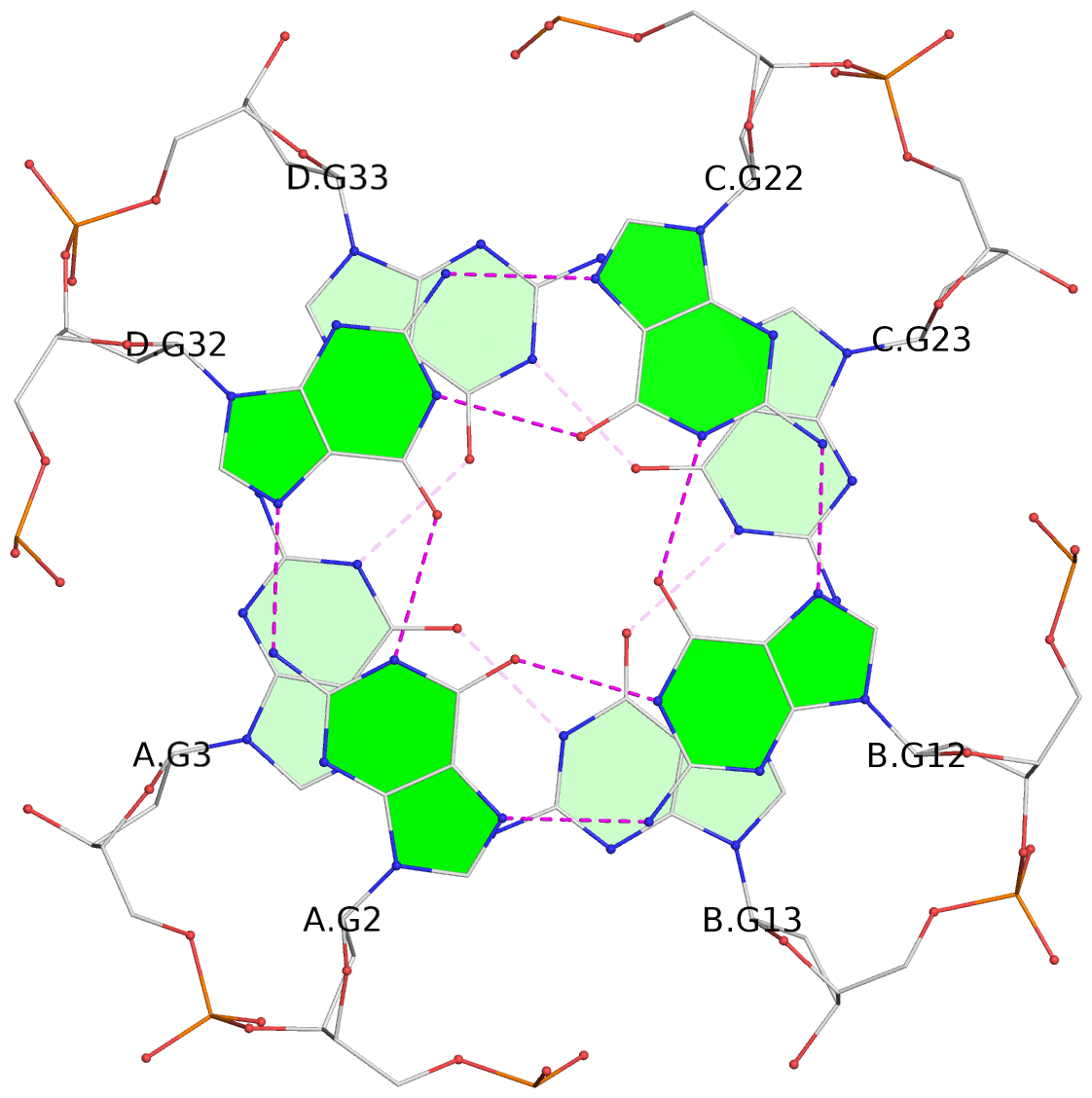
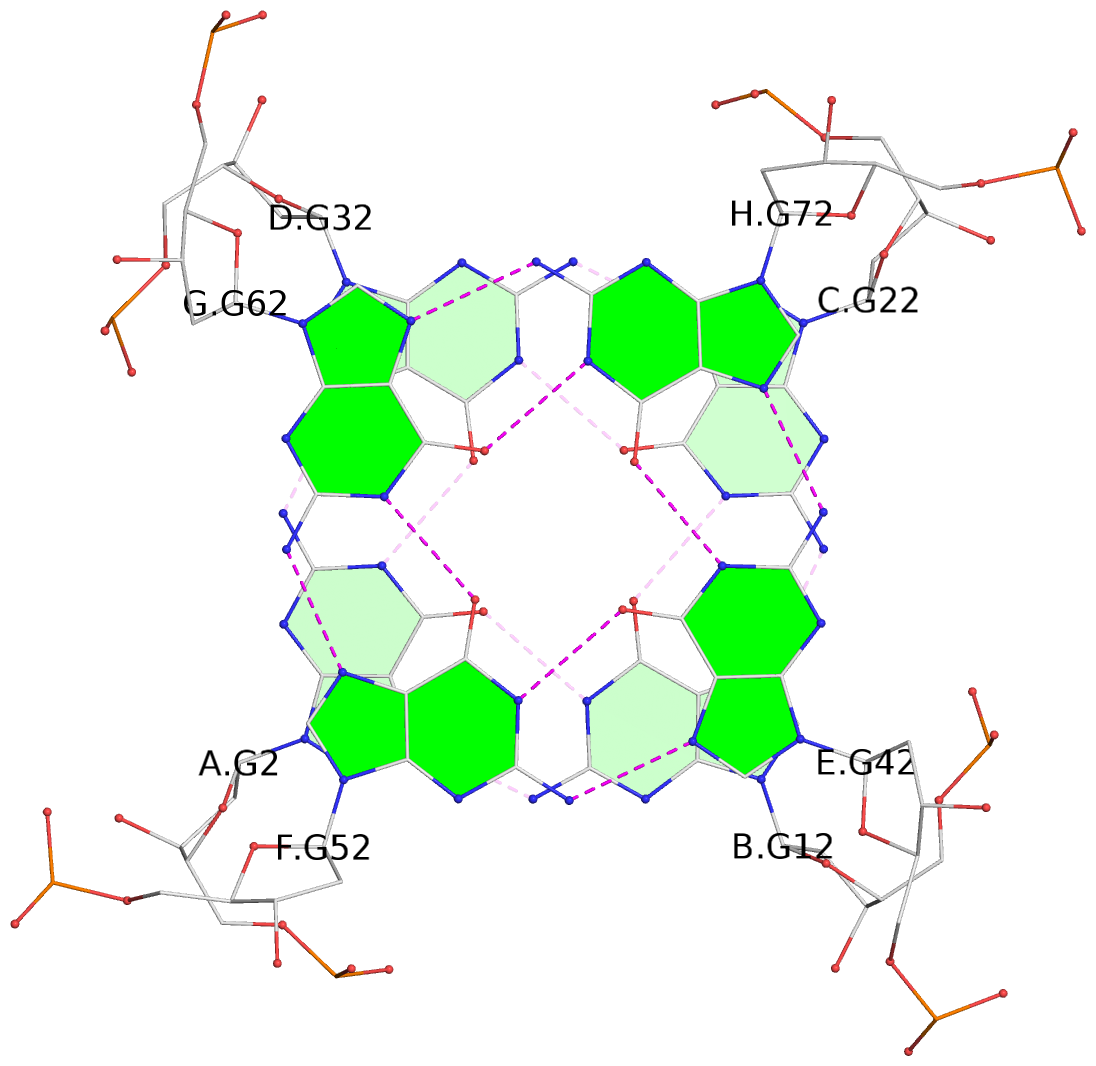
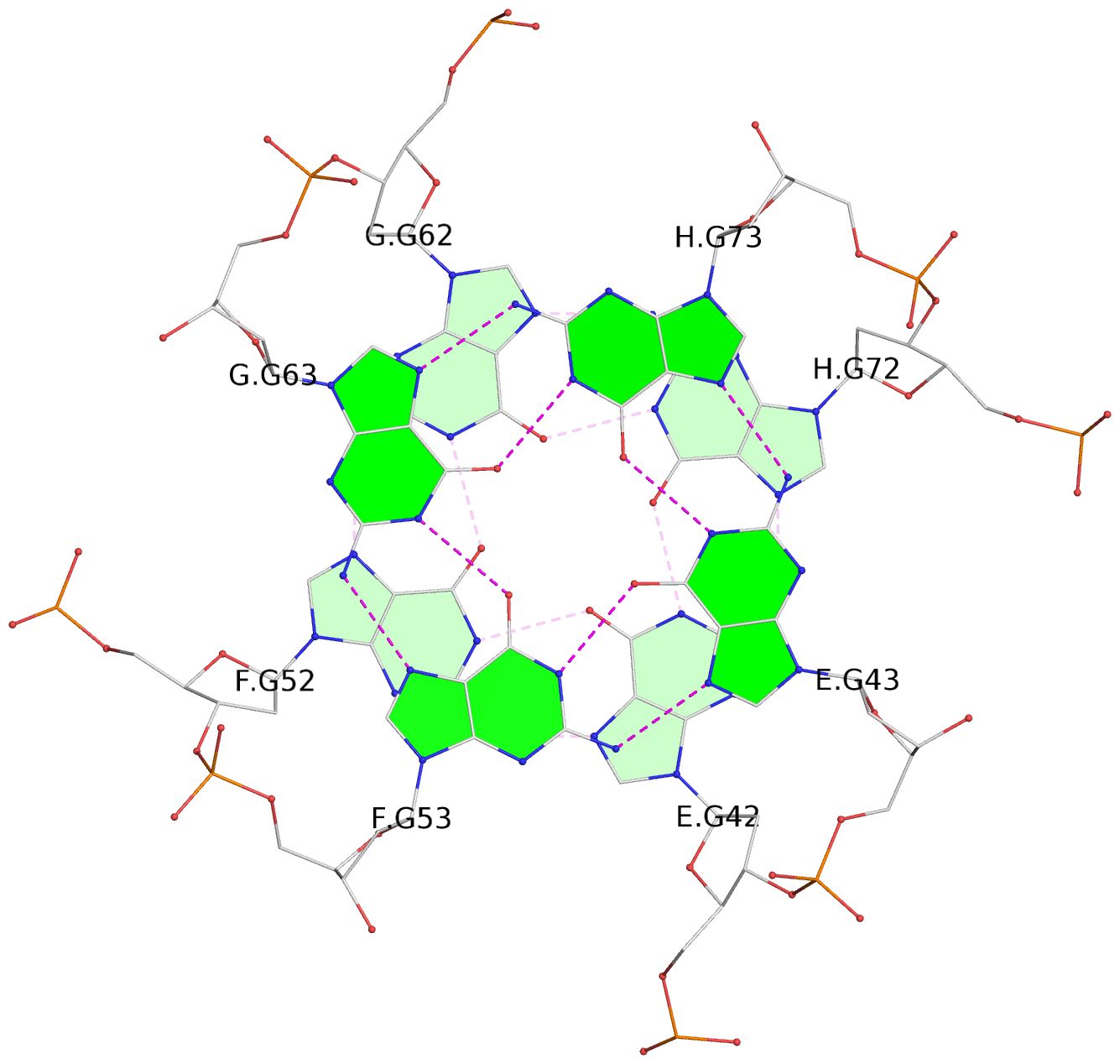
List of 16 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.214 type=bowl nts=4 GGGG A.DG2,D.DG32,C.DG22,B.DG12 2 glyco-bond=---- sugar=---- groove=---- planarity=0.200 type=bowl nts=4 GGGG A.DG3,D.DG33,C.DG23,B.DG13 3 glyco-bond=---- sugar=---- groove=---- planarity=0.237 type=bowl nts=4 GGGG A.DG4,D.DG34,C.DG24,B.DG14 4 glyco-bond=---- sugar=---- groove=---- planarity=0.289 type=bowl nts=4 GGGG A.DG5,D.DG35,C.DG25,B.DG15 5 glyco-bond=---- sugar=333. groove=---- planarity=0.041 type=planar nts=4 GGGG E.DG42,H.DG72,G.DG62,F.DG52 6 glyco-bond=---- sugar=---- groove=---- planarity=0.216 type=bowl nts=4 GGGG E.DG43,H.DG73,G.DG63,F.DG53 7 glyco-bond=---- sugar=---- groove=---- planarity=0.246 type=bowl nts=4 GGGG E.DG44,H.DG74,G.DG64,F.DG54 8 glyco-bond=---- sugar=---- groove=---- planarity=0.255 type=bowl nts=4 GGGG E.DG45,H.DG75,G.DG65,F.DG55 9 glyco-bond=---- sugar=3333 groove=---- planarity=0.045 type=planar nts=4 GGGG I.DG82,L.DG112,K.DG102,J.DG92 10 glyco-bond=---- sugar=---- groove=---- planarity=0.228 type=bowl nts=4 GGGG I.DG83,L.DG113,K.DG103,J.DG93 11 glyco-bond=---- sugar=---- groove=---- planarity=0.238 type=bowl nts=4 GGGG I.DG84,L.DG114,K.DG104,J.DG94 12 glyco-bond=---- sugar=---- groove=---- planarity=0.236 type=bowl nts=4 GGGG I.DG85,L.DG115,K.DG105,J.DG95 13 glyco-bond=---- sugar=---- groove=---- planarity=0.218 type=bowl nts=4 GGGG M.DG122,P.DG152,O.DG142,N.DG132 14 glyco-bond=---- sugar=---- groove=---- planarity=0.199 type=bowl nts=4 GGGG M.DG123,P.DG153,O.DG143,N.DG133 15 glyco-bond=---- sugar=---- groove=---- planarity=0.220 type=bowl nts=4 GGGG M.DG124,P.DG154,O.DG144,N.DG134 16 glyco-bond=---- sugar=---- groove=---- planarity=0.283 type=bowl nts=4 GGGG M.DG125,P.DG155,O.DG145,N.DG135
List of 2 G4-helices
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 8 G-tetrad layers, inter-molecular, with 2 stems
Helix#2, 8 G-tetrad layers, inter-molecular, with 2 stems
List of 4 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 4 G-tetrad layers, 0 loops, inter-molecular, UUUU, parallel, parallel(4+0)
Stem#2, 4 G-tetrad layers, 0 loops, inter-molecular, UUUU, parallel, parallel(4+0)
Stem#3, 4 G-tetrad layers, 0 loops, inter-molecular, UUUU, parallel, parallel(4+0)
Stem#4, 4 G-tetrad layers, 0 loops, inter-molecular, UUUU, parallel, parallel(4+0)
List of 2 G4 coaxial stacks
1 G4 helix#1 contains 2 G4 stems: [#1,#2] [5'/5'] 2 G4 helix#2 contains 2 G4 stems: [#3,#4] [5'/5']