Detailed DSSR results for the G-quadruplex: PDB entry 2a5r
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 2a5r
- Class
- DNA
- Method
- NMR
- Summary
- Complex of tetra-(4-n-methylpyridyl) porphin with monomeric parallel-stranded DNA tetraplex, snap-back 3+1 3' g-tetrad, single-residue chain reversal loops, gag triad in the context of gaag diagonal loop, c-myc promoter, NMR, 6 struct.
- Reference
- Phan AT, Kuryavyi V, Gaw HY, Patel DJ (2005): "Small-molecule interaction with a five-guanine-tract G-quadruplex structure from the human MYC promoter." Nat.Chem.Biol., 1, 167-173. doi: 10.1038/nchembio723.
- Abstract
- It has been widely accepted that DNA can adopt other biologically relevant structures beside the Watson-Crick double helix. One recent important example is the guanine-quadruplex (G-quadruplex) structure formed by guanine tracts found in the MYC (or c-myc) promoter region, which regulates the transcription of the MYC oncogene. Stabilization of this G-quadruplex by ligands, such as the cationic porphyrin TMPyP4, decreases the transcriptional level of MYC. Here, we report the first structure of a DNA fragment containing five guanine tracts from this region. An unusual G-quadruplex fold, which was derived from NMR restraints using unambiguous model-independent resonance assignment approaches, involves a core of three stacked guanine tetrads formed by four parallel guanine tracts with all anti guanines and a snapback 3'-end syn guanine. We have determined the structure of the complex formed between this G-quadruplex and TMPyP4. This structural information, combined with details of small-molecule interaction, provides a platform for the design of anticancer drugs targeting multi-guanine-tract sequences that are found in the MYC and other oncogenic promoters, as well as in telomeres.
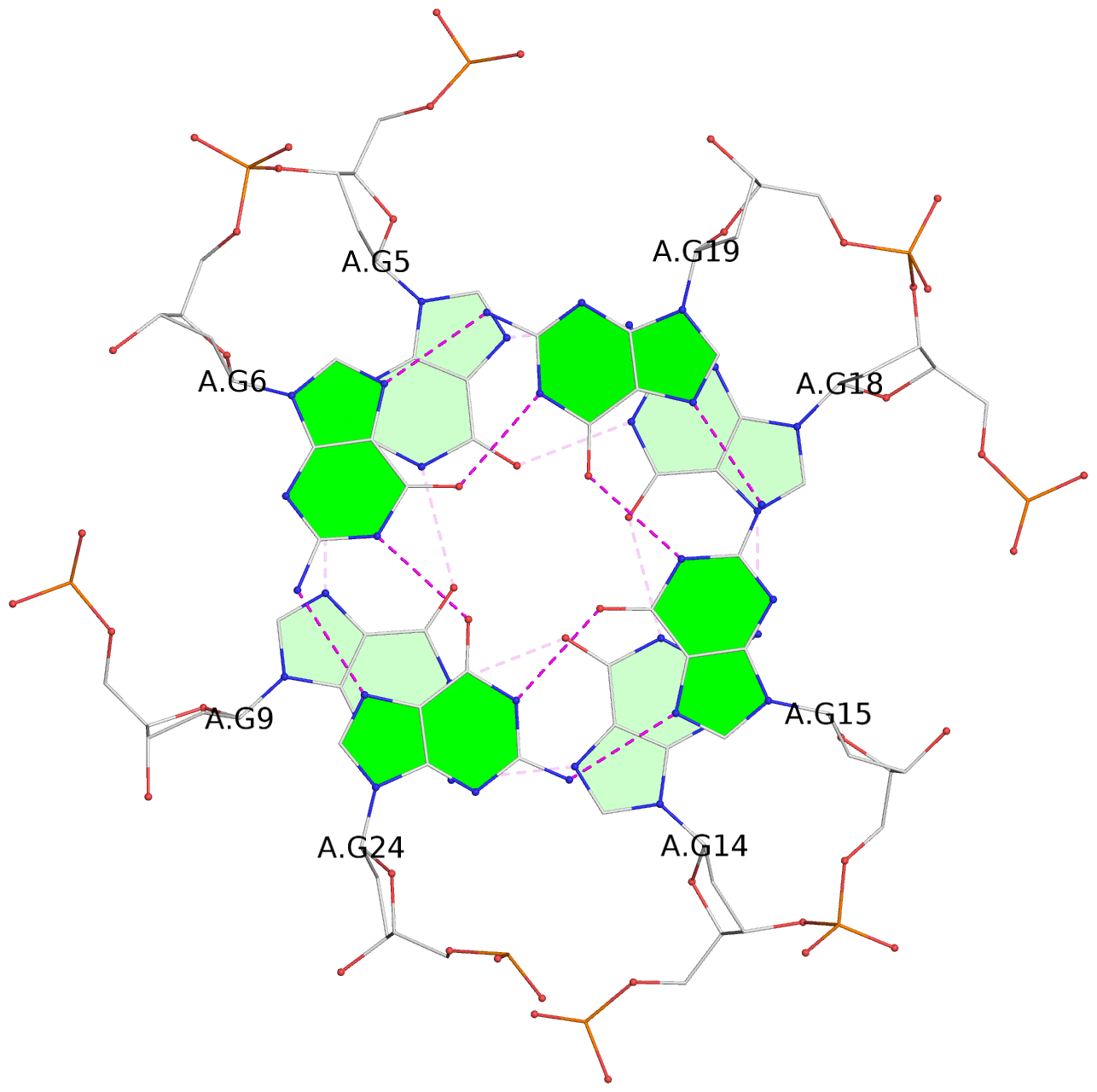
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 2(-P-P-P), parallel(4+0), UUUU
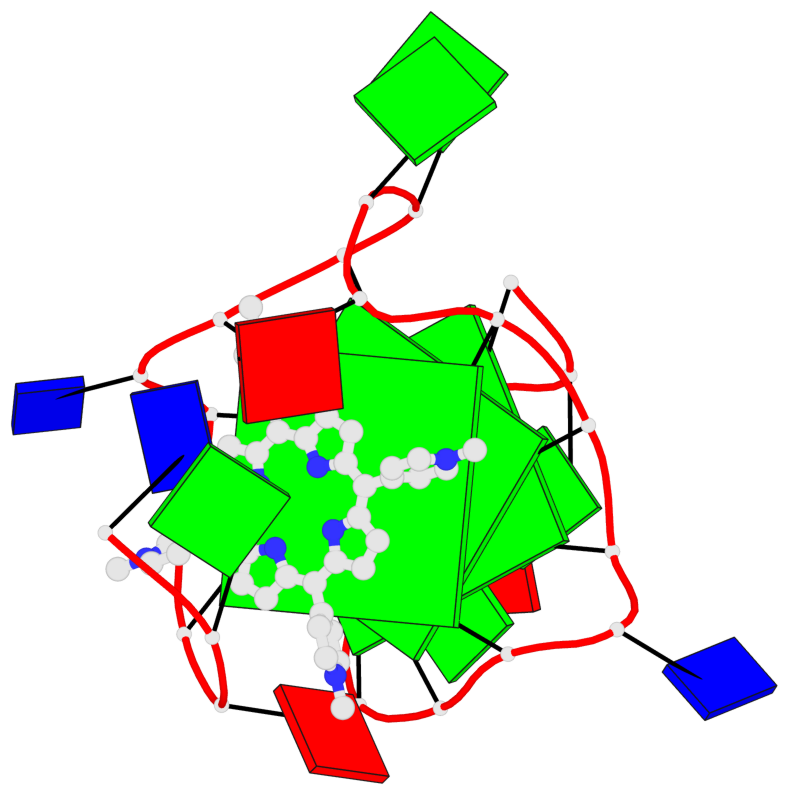
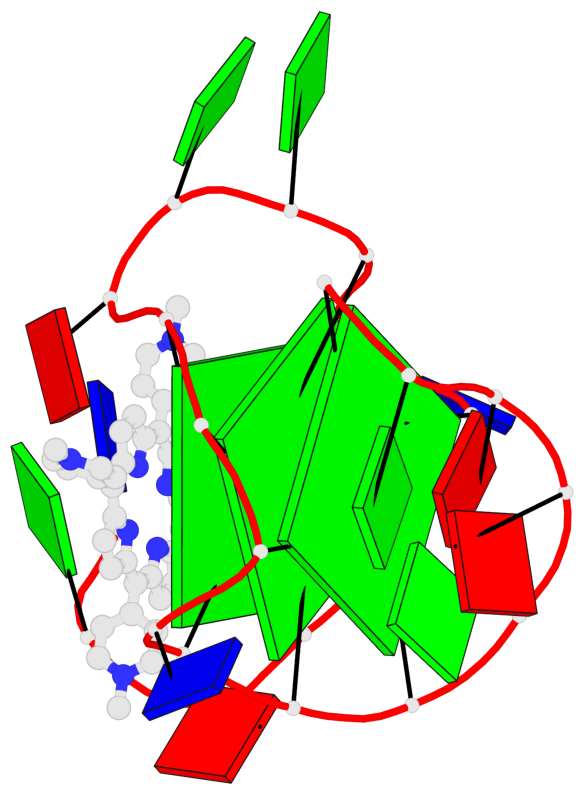
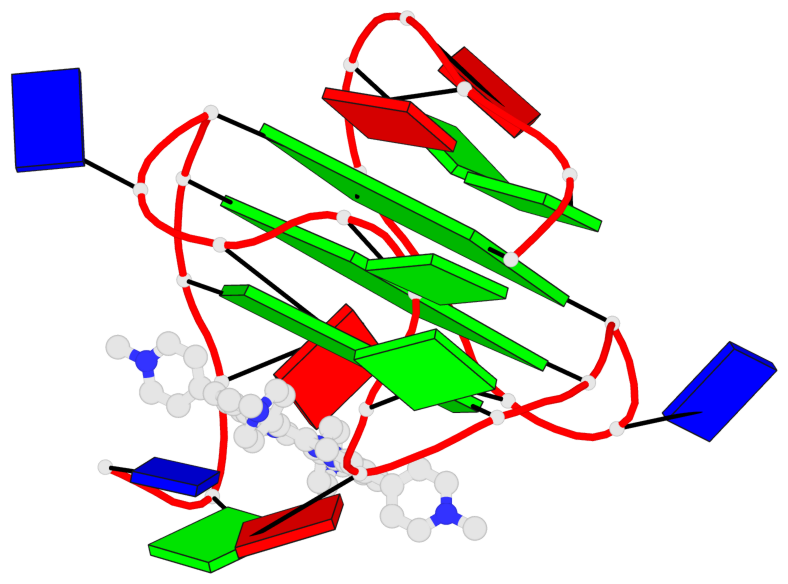
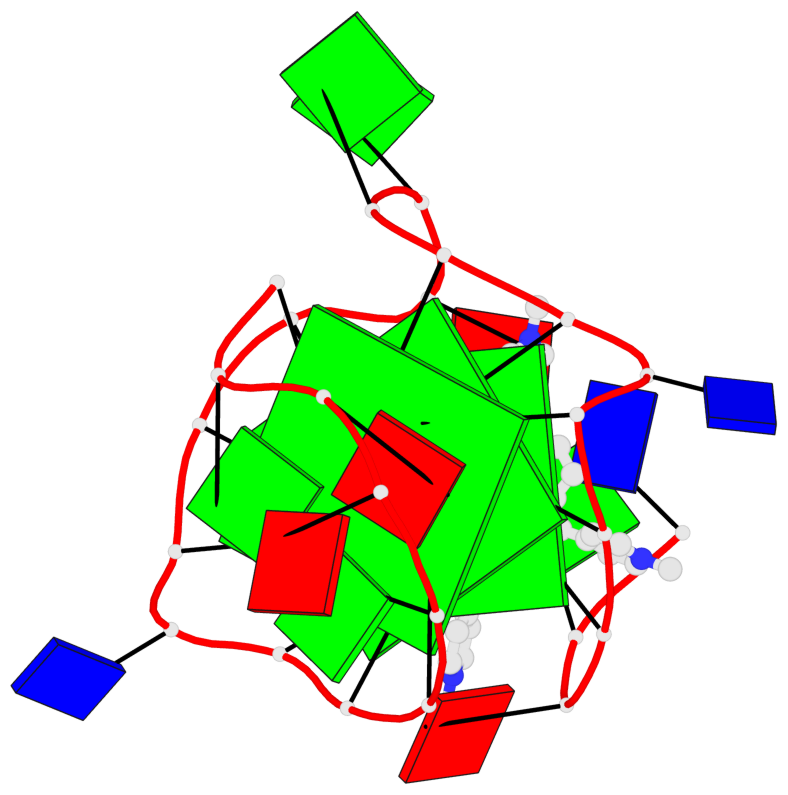
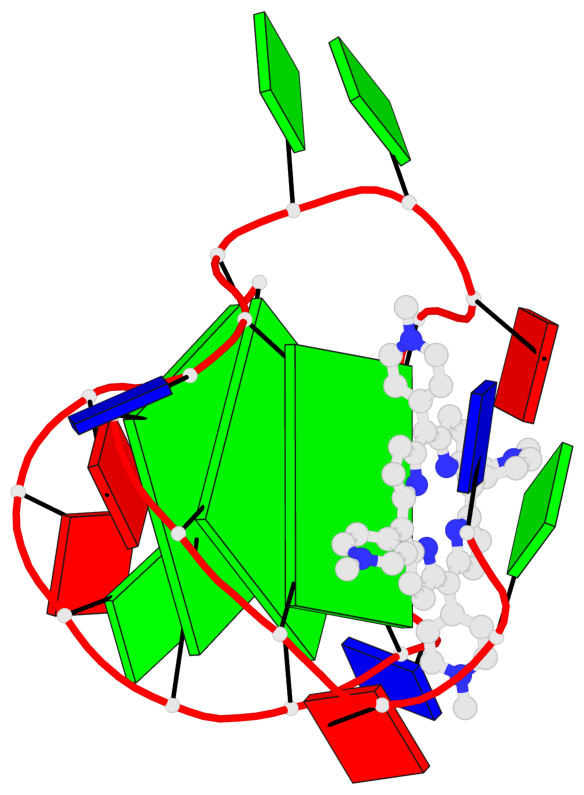
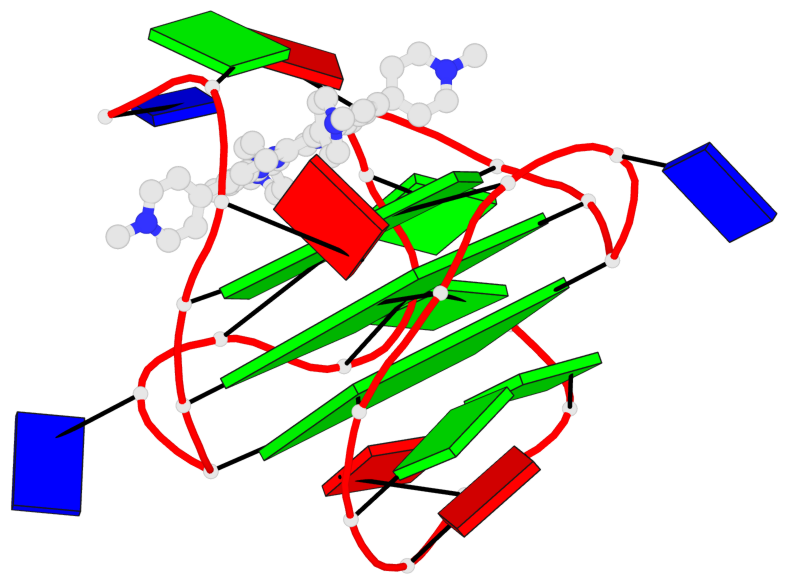
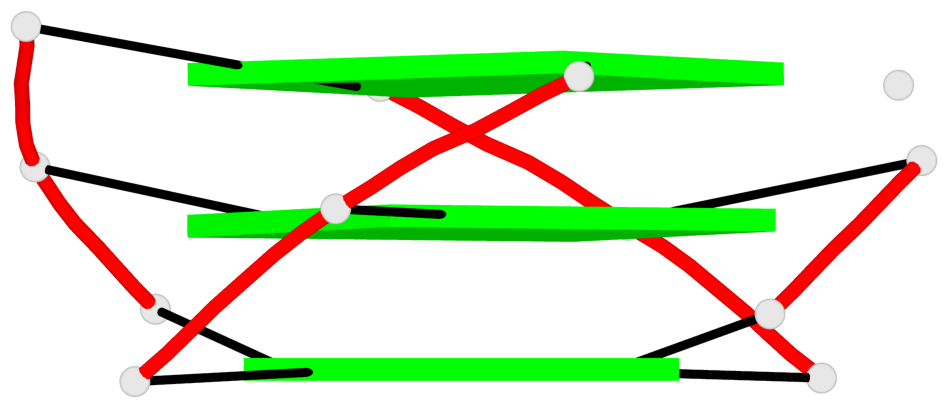
Base-block schematics in six views
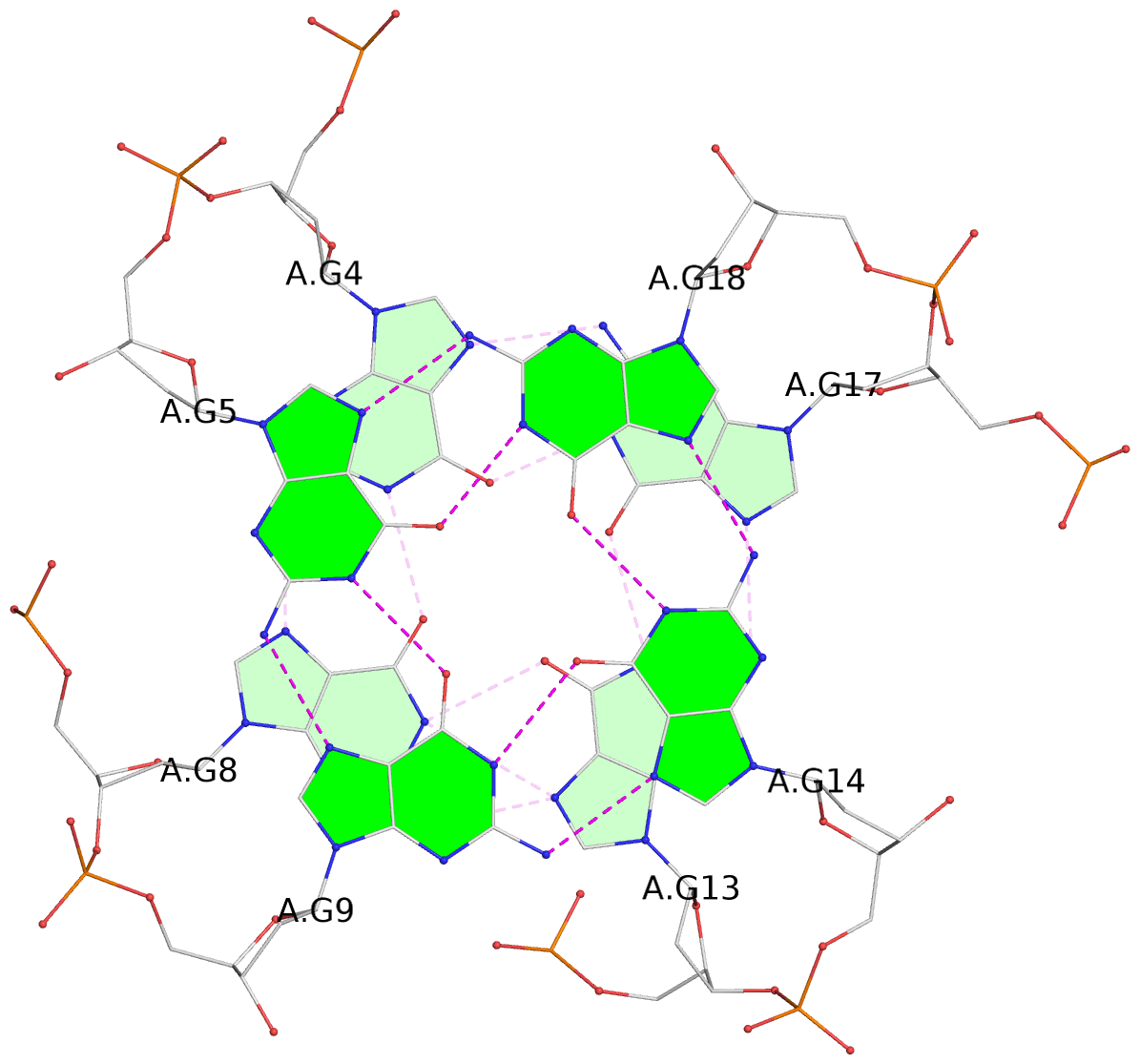
List of 3 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.114 type=planar nts=4 GGGG A.DG4,A.DG8,A.DG13,A.DG17 2 glyco-bond=---- sugar=.--- groove=---- planarity=0.240 type=other nts=4 GGGG A.DG5,A.DG9,A.DG14,A.DG18 3 glyco-bond=-s-- sugar=---- groove=wn-- planarity=0.304 type=other nts=4 GGGG A.DG6,A.DG24,A.DG15,A.DG19
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 2 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 2(-P-P-P), parallel(4+0)
List of 1 non-stem G4-loop (including the two closing Gs)
1 type=diagonal helix=#1 nts=6 GGAAGG A.DG19,A.DG20,A.DA21,A.DA22,A.DG23,A.DG24