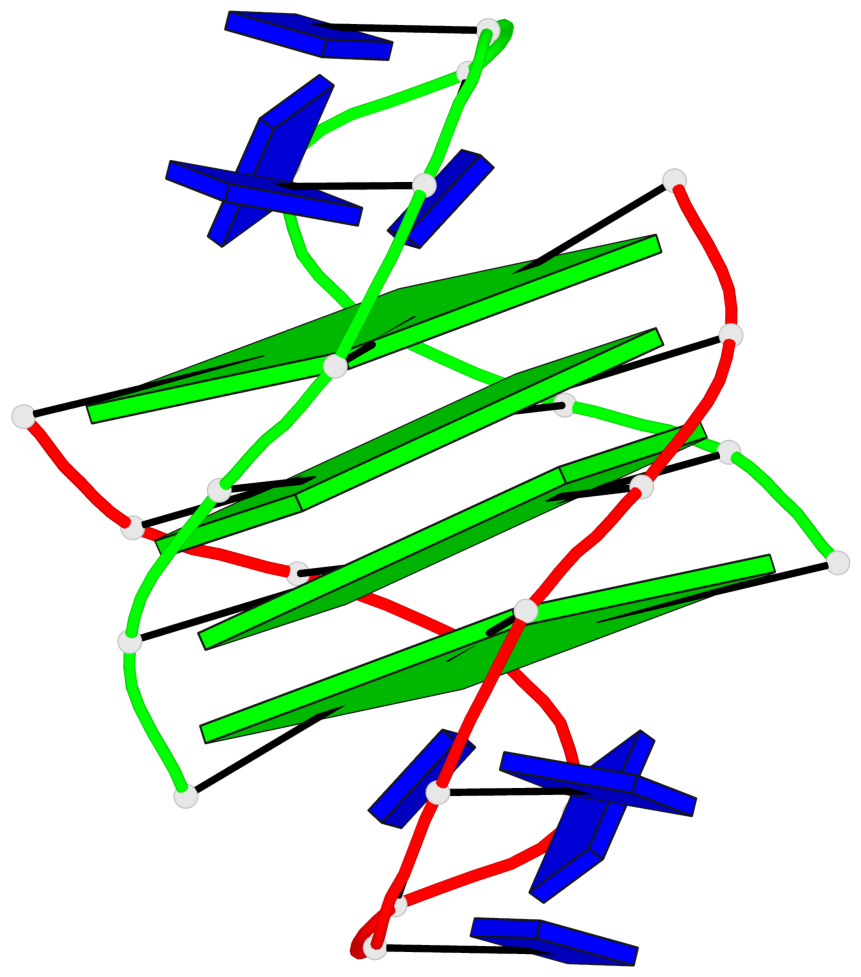
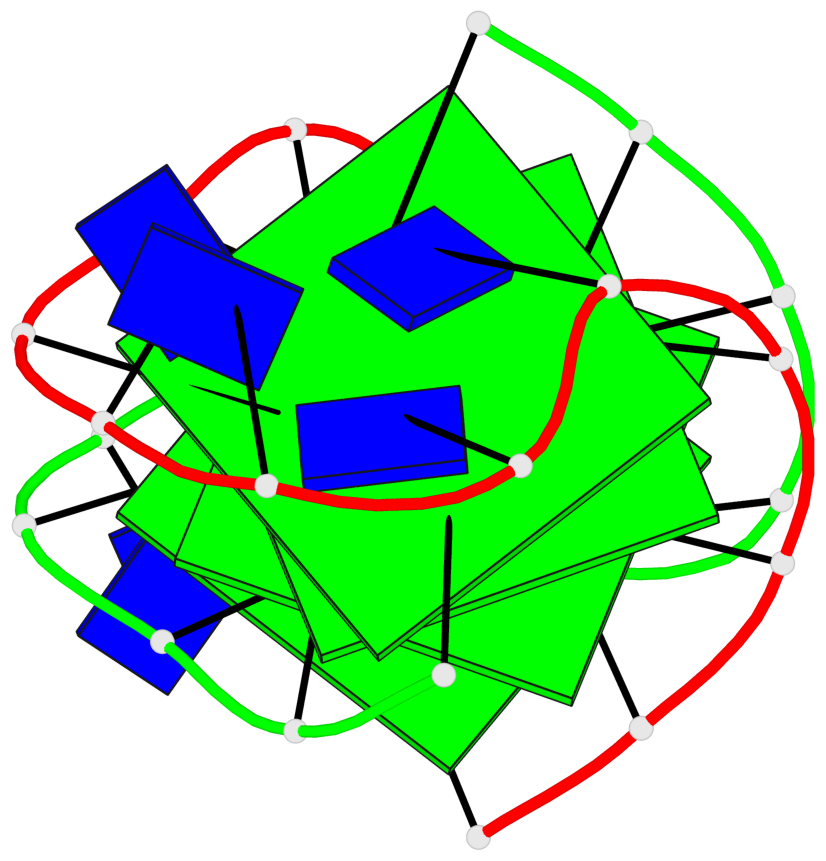
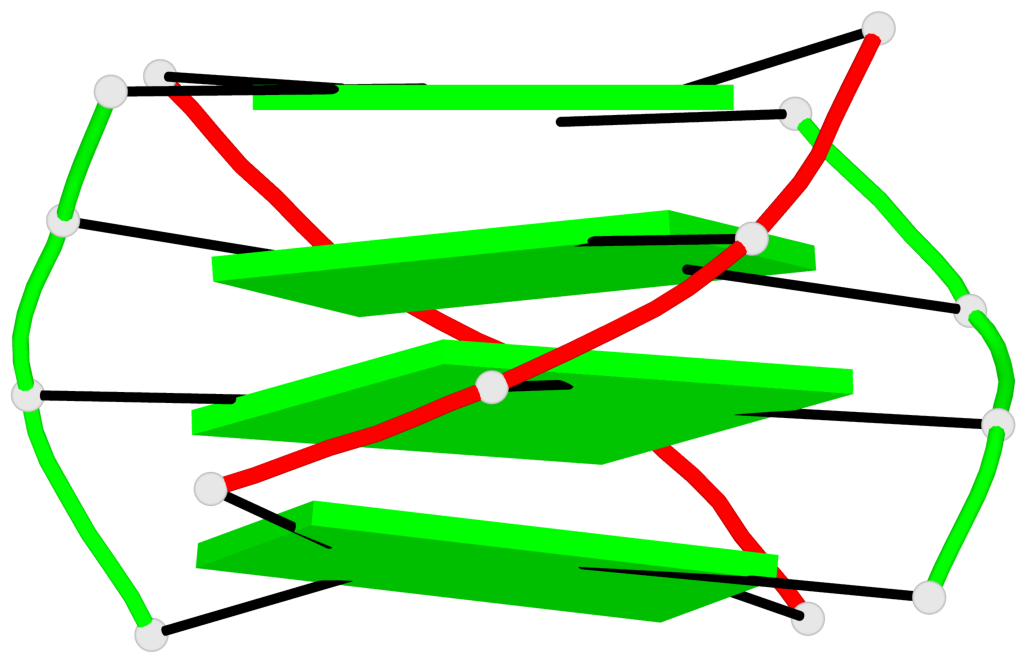
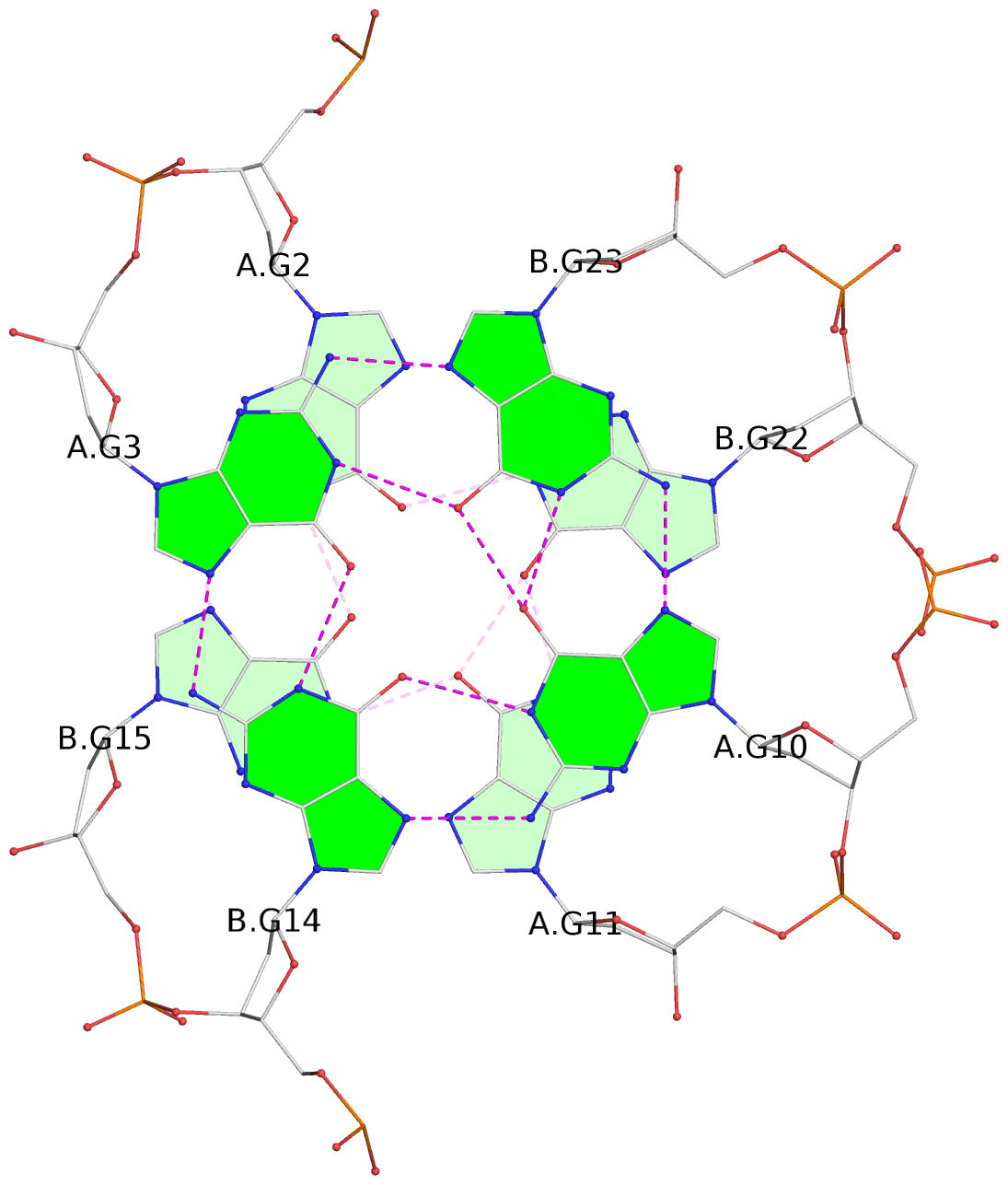
Detailed DSSR results for the G-quadruplex: PDB entry 2akg
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 2akg
- Class
- DNA
- Method
- NMR
- Summary
- Thallium form of the g-quadruplex from oxytricha nova, d(g4t4g4)2
- Reference
- Gill ML, Strobel SA, Loria JP (2005): "(205)Tl NMR methods for the characterization of monovalent cation binding to nucleic acids." J.Am.Chem.Soc., 127, 16723-16732. doi: 10.1021/ja055358f.
- Abstract
- Monovalent cations play an important role in many biological functions. The guanine rich sequence, d(G4T4G4), requires monovalent cations for formation of the G-quadruplex, d(G4T4G4)2. This requirement can be satisfied by thallium (Tl+), a potassium (K+) surrogate. To verify that the structure of d(G4T4G4)2 in the presence of Tl+ is similar to the K+-form of the G-quadruplex, the solution structure of the Tl+-form of d(G4T4G4)2 was determined. The 10 lowest energy structures have an all atom RMSD of 0.76 +/- 0.16 A. Comparison of this structure to the identical G-quadruplex formed in the presence of K+ validates the isomorphous nature of Tl+ and K+. Using a 1H-205Tl spin-echo difference experiment we show that, in the Tl+-form of d(G4T4G4)2, small scalar couplings (<1 Hz) exist between 205Tl and protons in the G-quadruplex. These data comprise the first 1H-205Tl scalar couplings observed in a biological system and have the potential to provide important constraints for structure determination. These experiments can be applied to any system in which the substituted Tl+ cations are in slow exchange with the bulk ions in solution.
- G4 notes
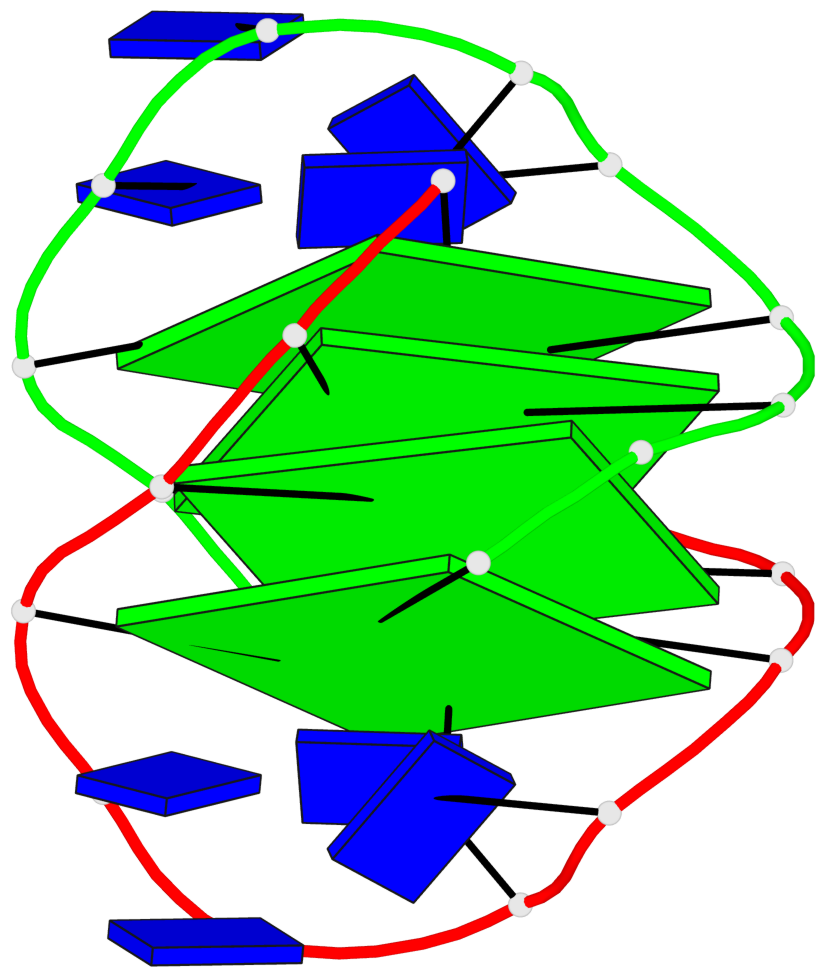
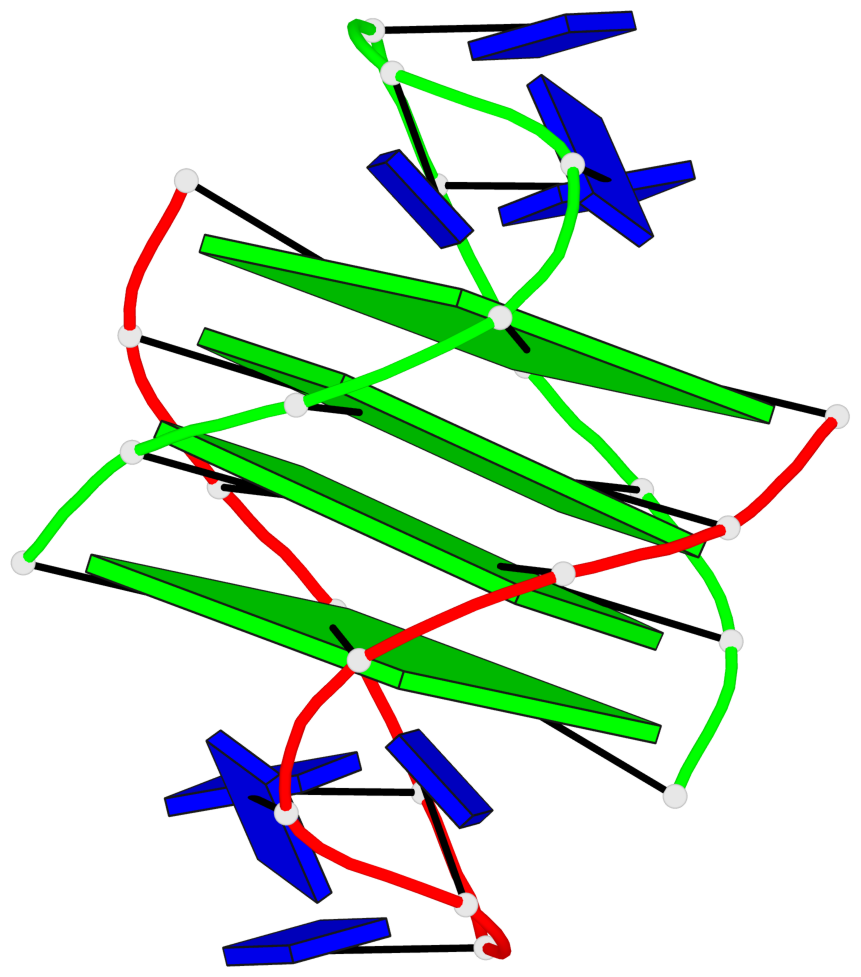
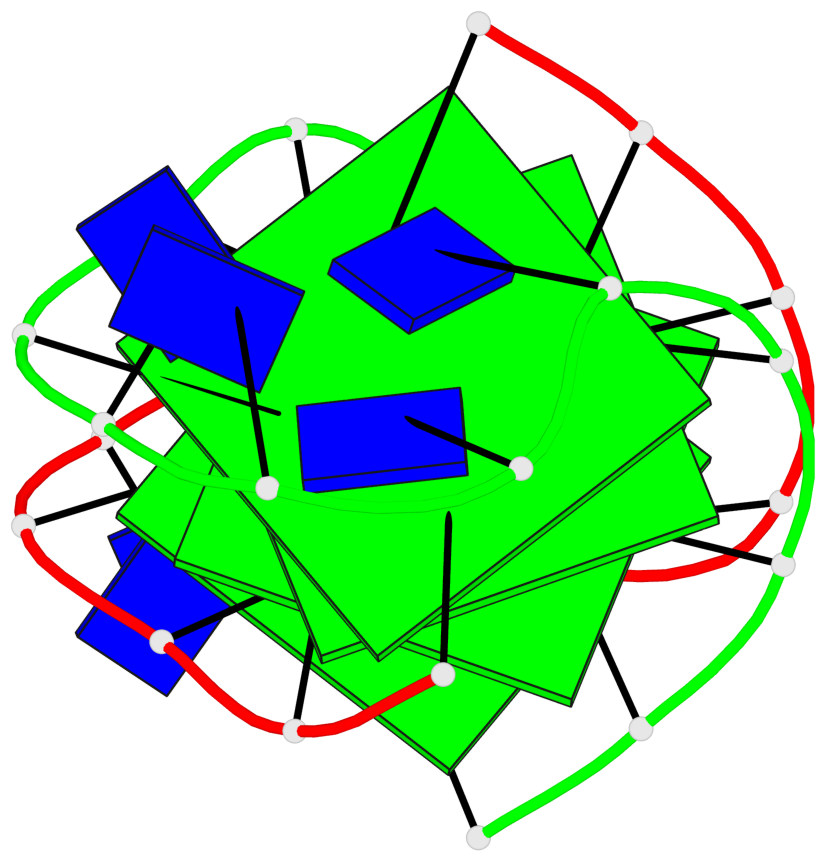
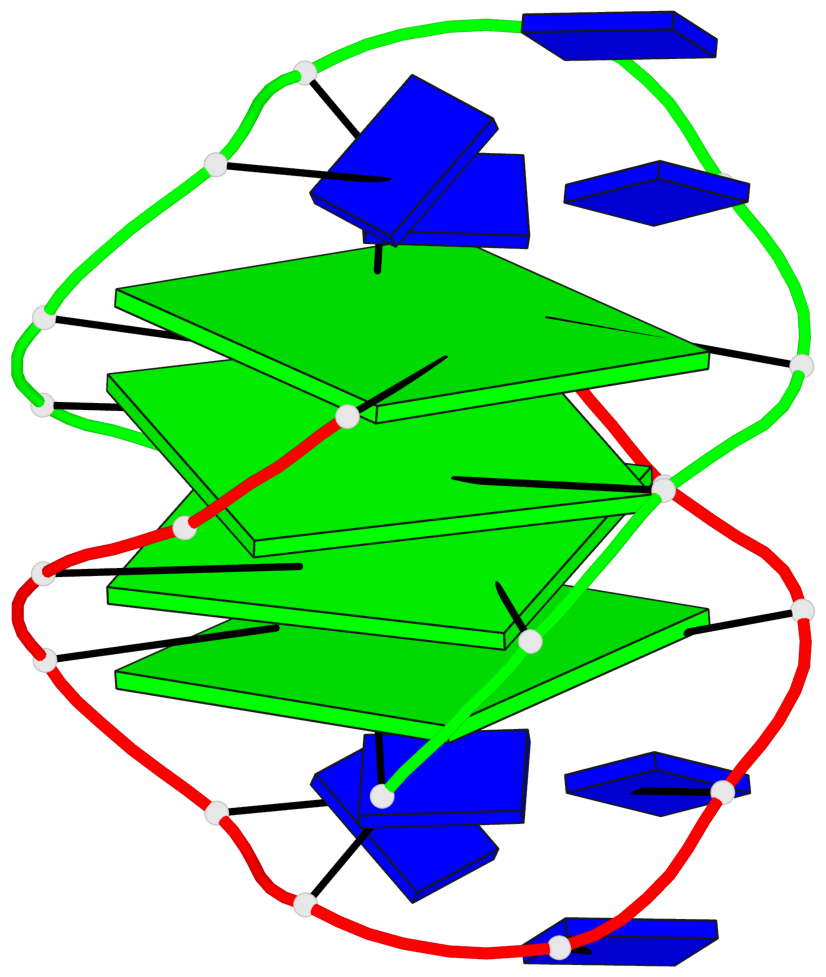
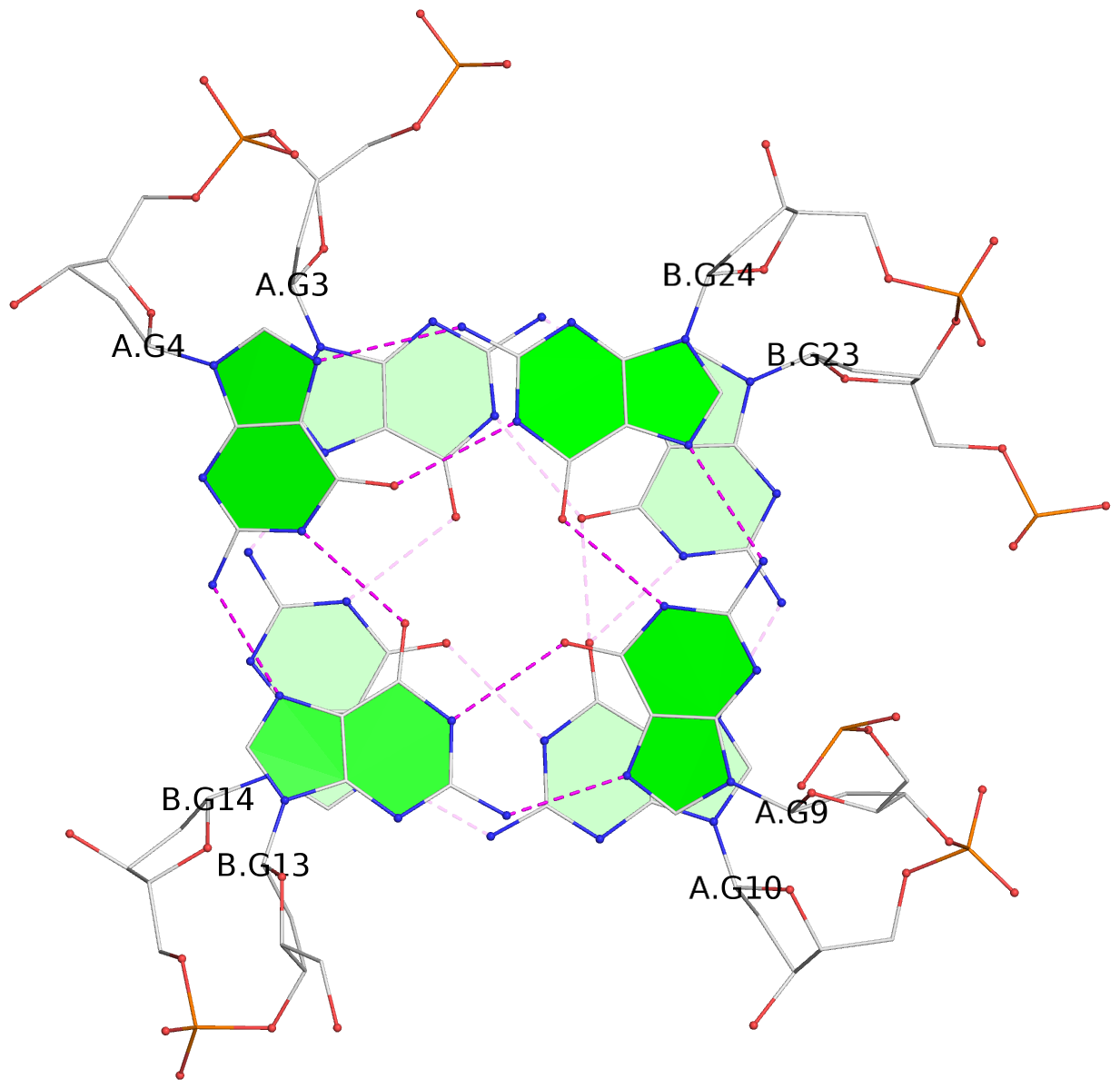
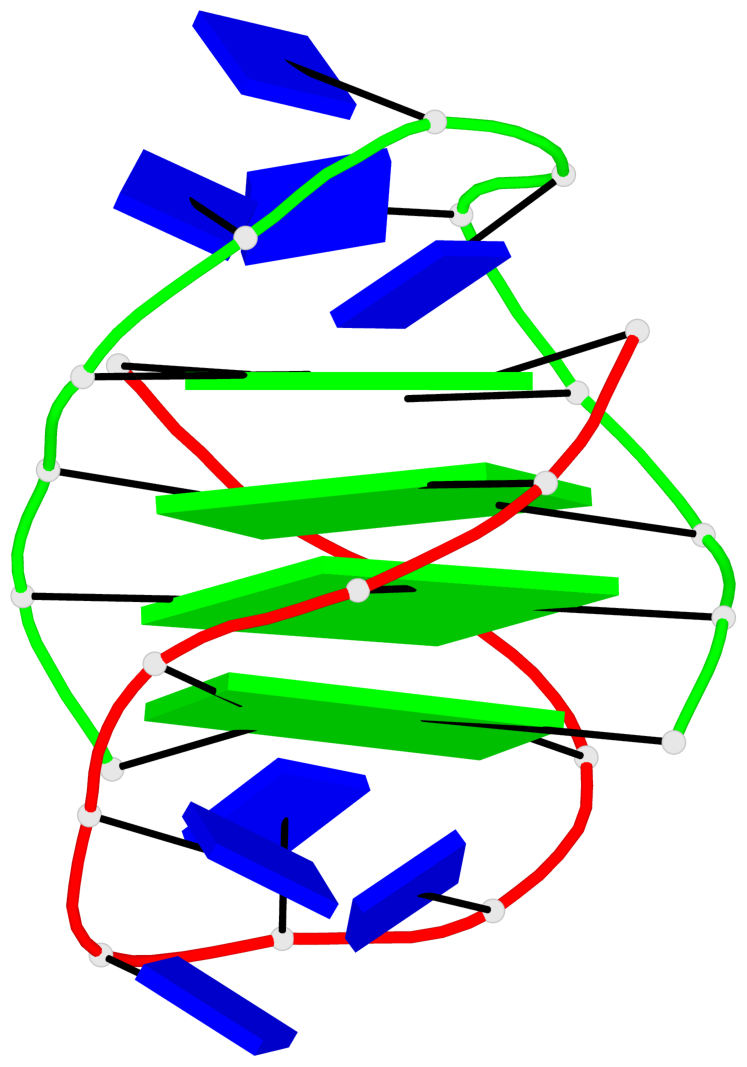
- 4 G-tetrads, 1 G4 helix, 1 G4 stem, (2+2), UDDU
Base-block schematics in six views
List of 4 G-tetrads
1 glyco-bond=s--s sugar=.-.. groove=w-n- planarity=0.339 type=other nts=4 GGGG A.DG1,B.DG16,A.DG12,B.DG21 2 glyco-bond=-ss- sugar=-.-- groove=w-n- planarity=0.098 type=planar nts=4 GGGG A.DG2,B.DG15,A.DG11,B.DG22 3 glyco-bond=s--s sugar=.--- groove=w-n- planarity=0.094 type=planar nts=4 GGGG A.DG3,B.DG14,A.DG10,B.DG23 4 glyco-bond=-ss- sugar=-... groove=w-n- planarity=0.335 type=other nts=4 GGGG A.DG4,B.DG13,A.DG9,B.DG24
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 4 G-tetrad layers, inter-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.