Detailed DSSR results for the G-quadruplex: PDB entry 2avj
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 2avj
- Class
- DNA
- Method
- X-ray (2.39 Å)
- Summary
- G4(br)uttg4 dimeric quadruplex
- Reference
- Hazel P, Parkinson GN, Neidle S (2006): "Topology variation and loop structural homology in crystal and simulated structures of a bimolecular DNA quadruplex." J.Am.Chem.Soc., 128, 5480-5487. doi: 10.1021/ja058577+.
- Abstract
- The topology of DNA quadruplexes depends on the nature and number of the nucleotides linking G-quartet motifs. To assess the effects of a three-nucleotide TTT linker, the crystal structure of the DNA sequence d(G(4)T(3)G(4)) has been determined at 1.5 A resolution, together with that of the brominated analogue d(G(4)(Br)UTTG(4)) at 2.4 A resolution. Both sequences form bimolecular intermolecular G-quadruplexes with lateral loops. d(G(4)(Br)UTTG(4)) crystallized in the monoclinic space group P2(1) with three quadruplex molecules in the asymmetric unit, two associating together as a head-to-head stacked dimer, and the third as a single head-to-tail dimer. The head-to-head dimers have two lateral loops on the same G-quadruplex face and form an eight-G-quartet stack, with a linear array of seven K(+) ions between the quartets. d(G(4)T(3)G(4)) crystallized in the orthorhombic space group C222 and has a structure very similar to the head-to-tail dimer in the P2(1) unit cell. The sequence studied here is able to form several different folds; however, all four quadruplexes in the two structures have lateral loops, in contrast to the diagonal loops reported for the analogous quadruplex with T(4) loops. A total of seven independent T(3) loops were observed in the two structures. These can be classified into two discrete conformational classes, suggesting that these represent preferred loop conformations that are independent of crystal-packing forces.
- G4 notes
- 12 G-tetrads, 2 G4 helices, 3 G4 stems, 1 G4 coaxial stack, (2+2), UDDU; (2+2), UDUD; (2+2), UUDD, coaxial interfaces: mixed
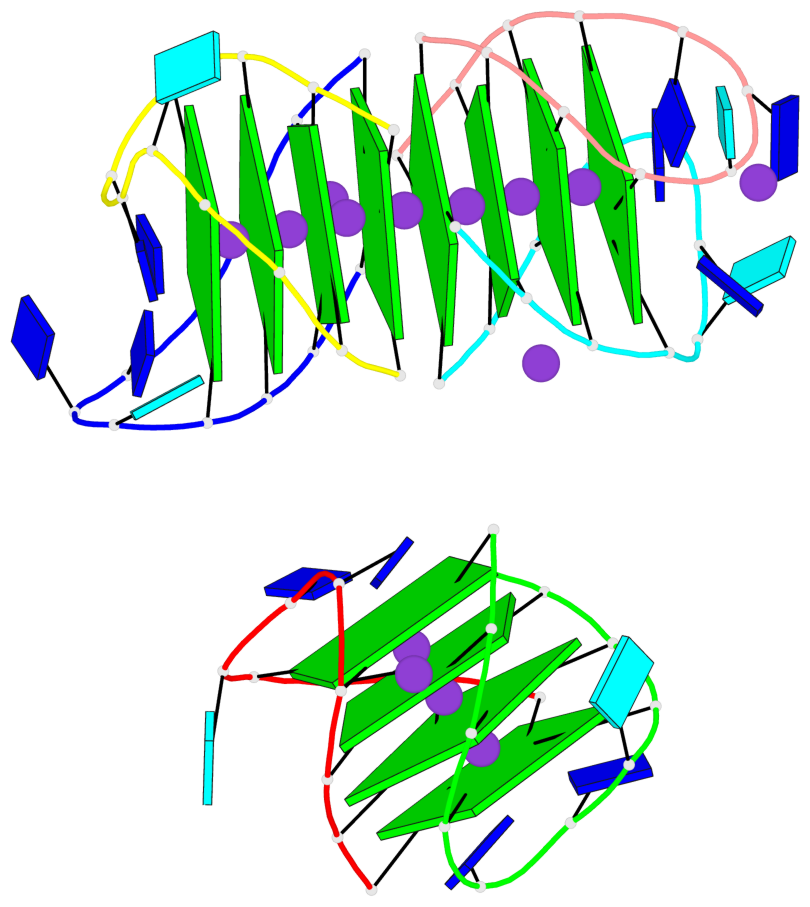
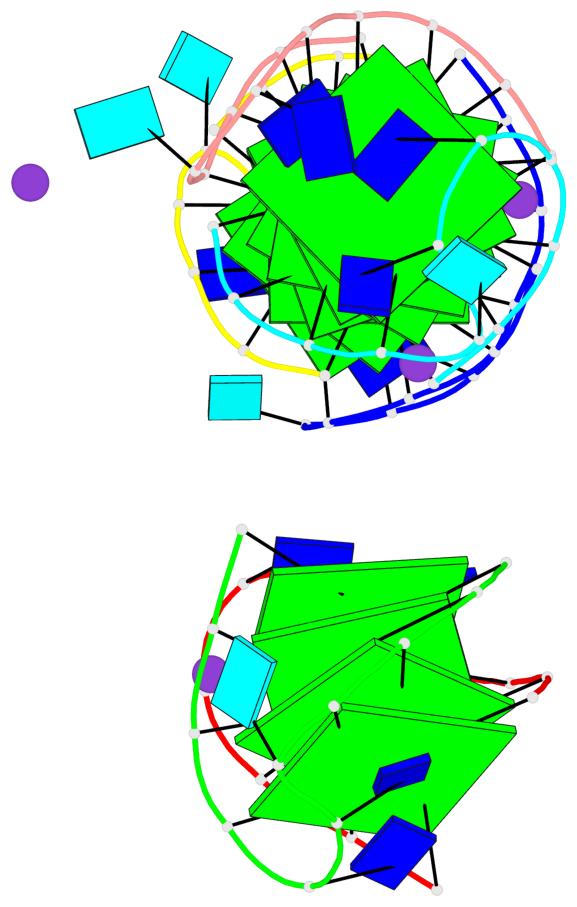
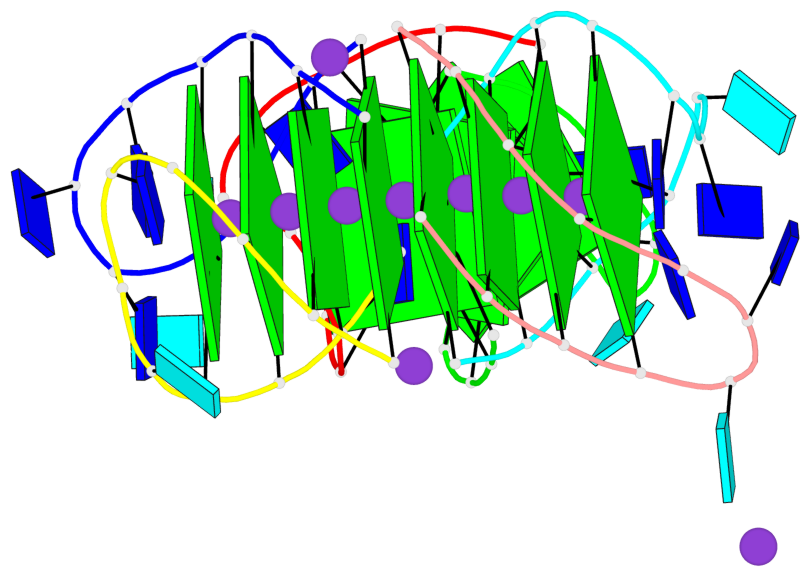
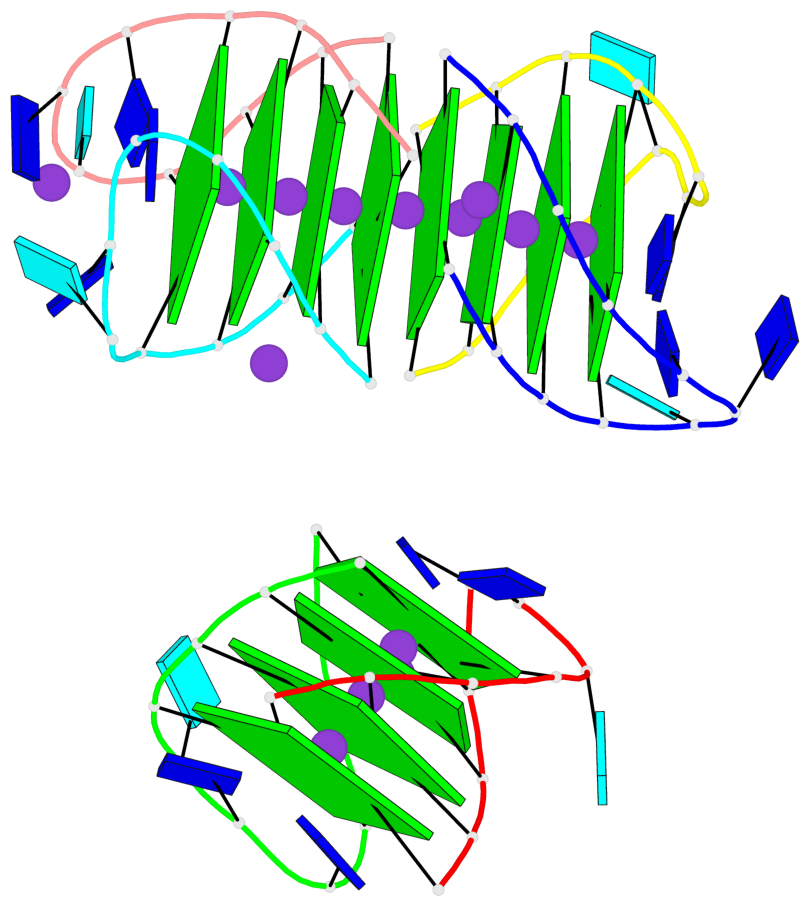
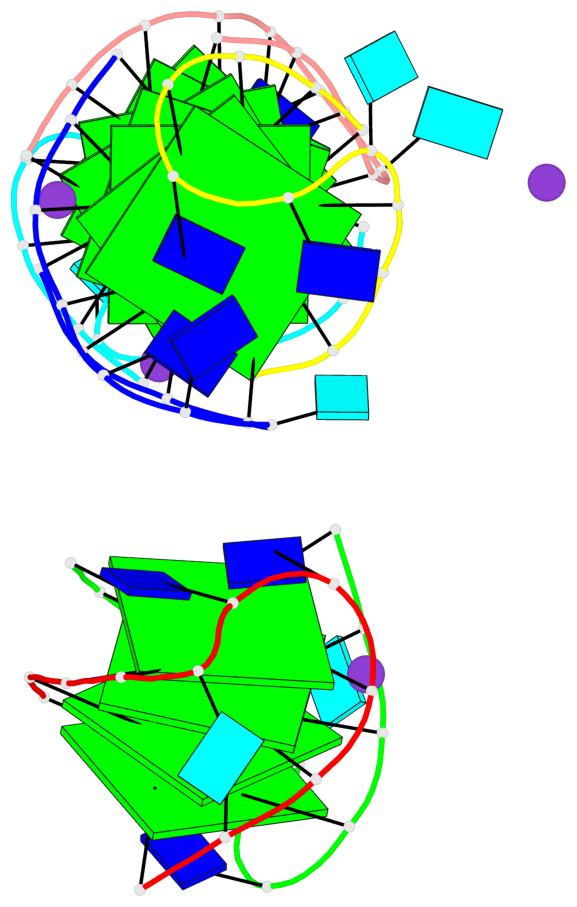
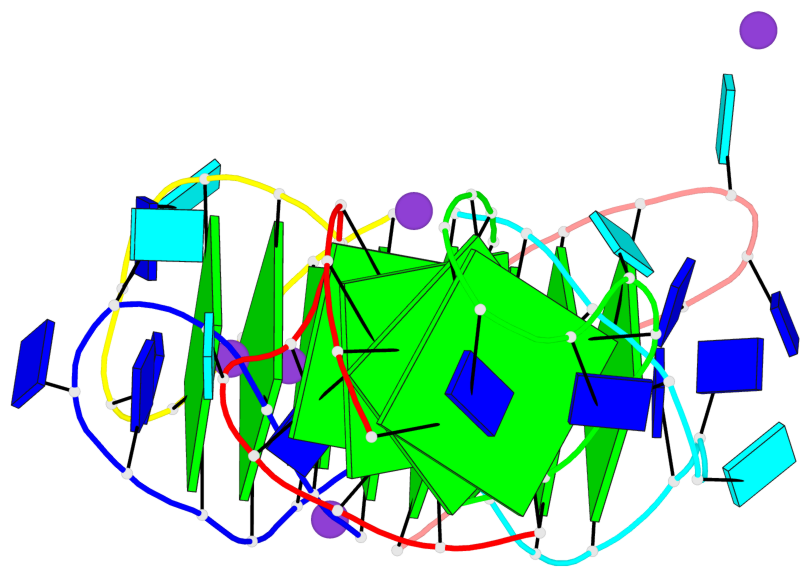
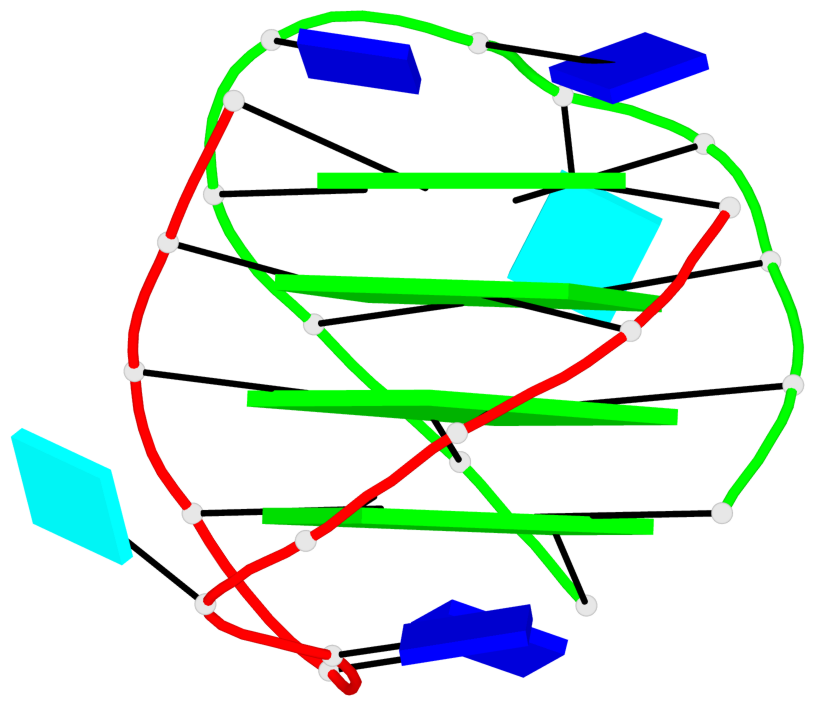
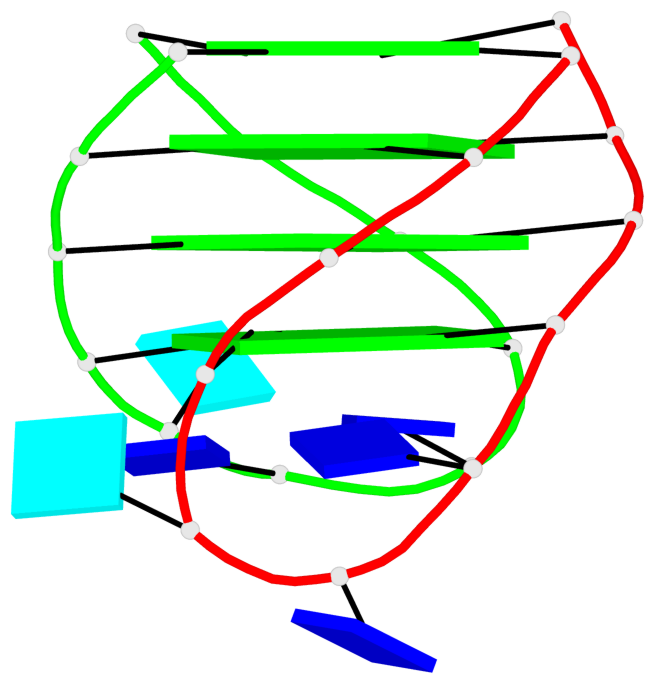
Base-block schematics in six views
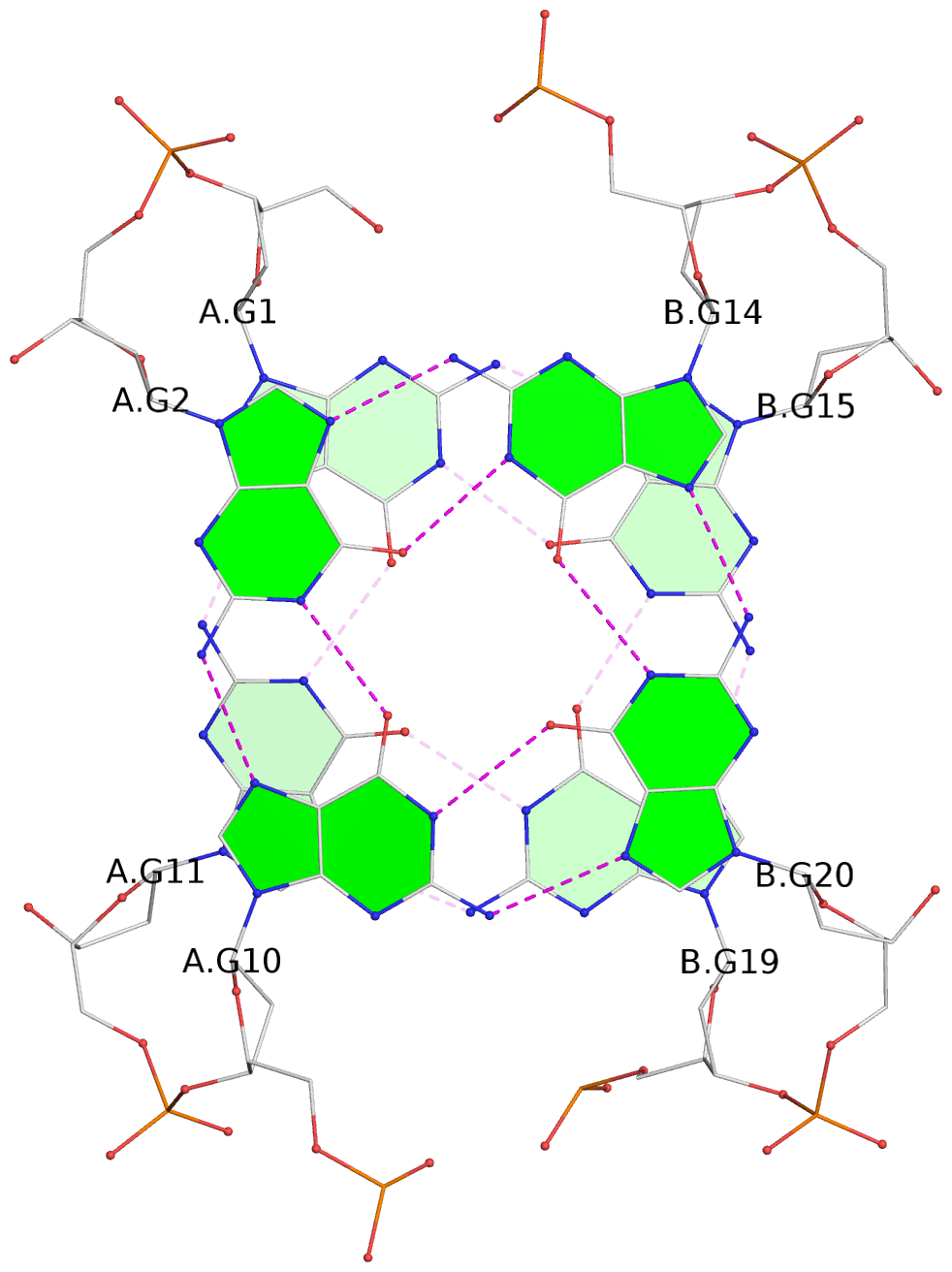
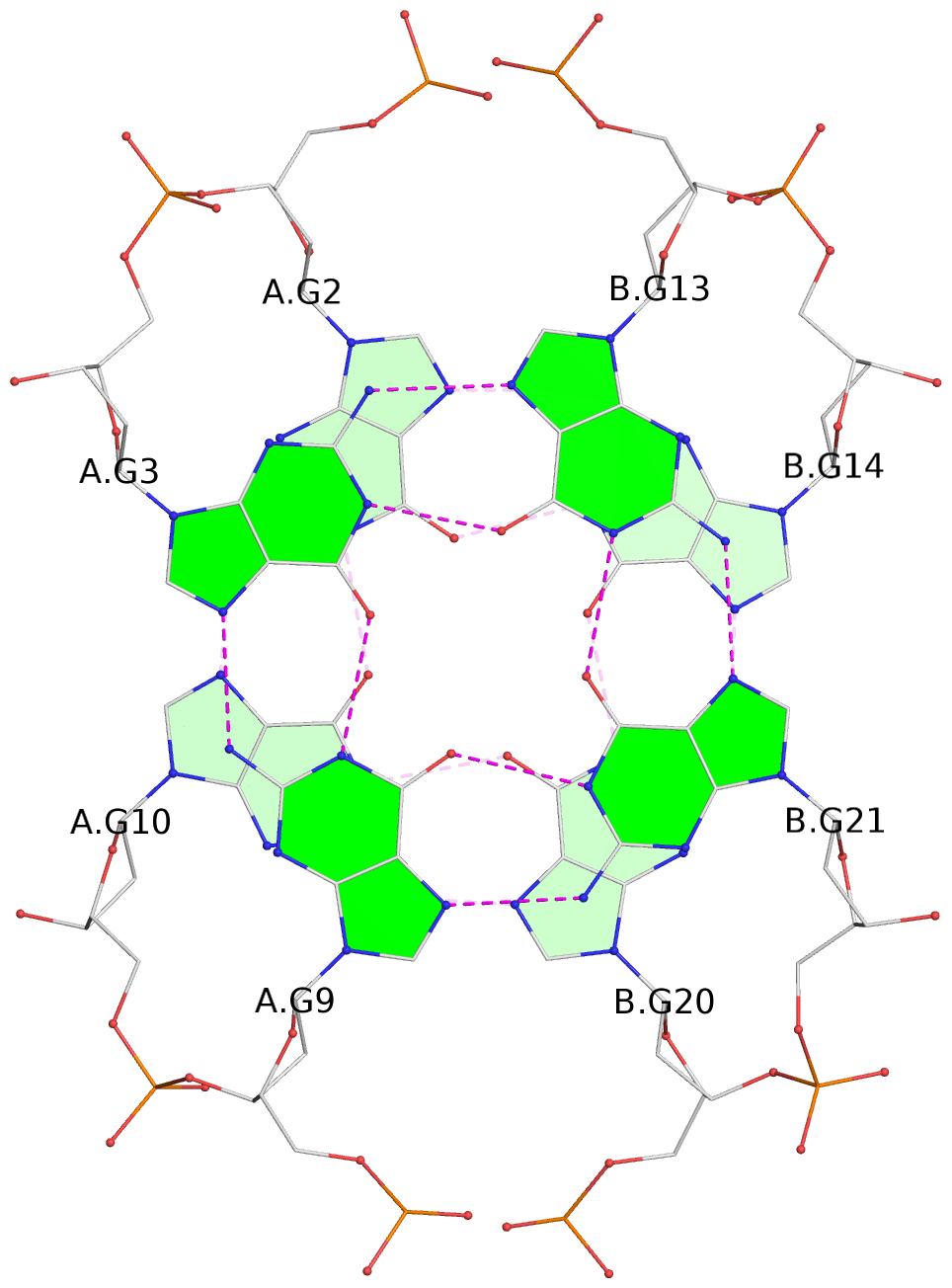
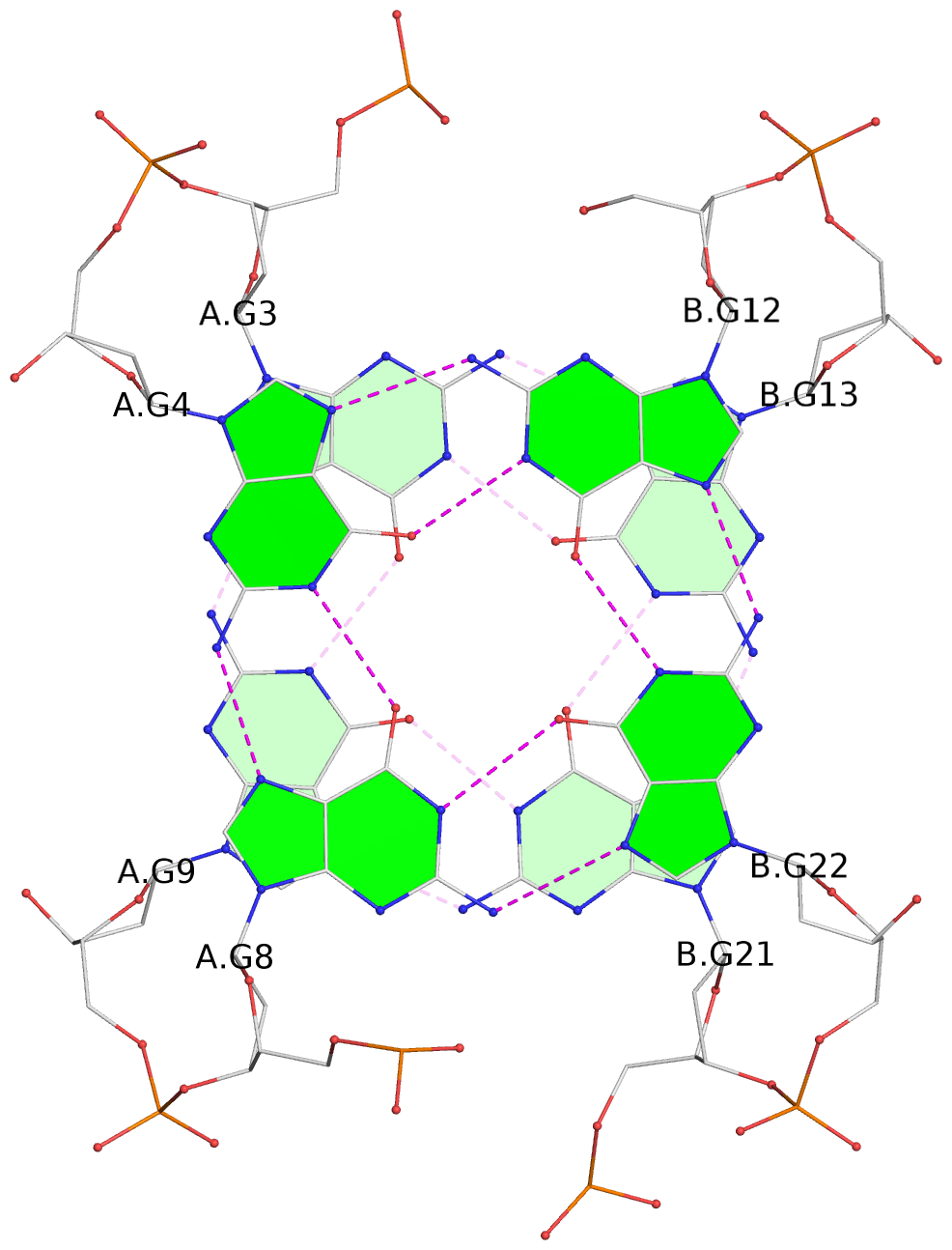
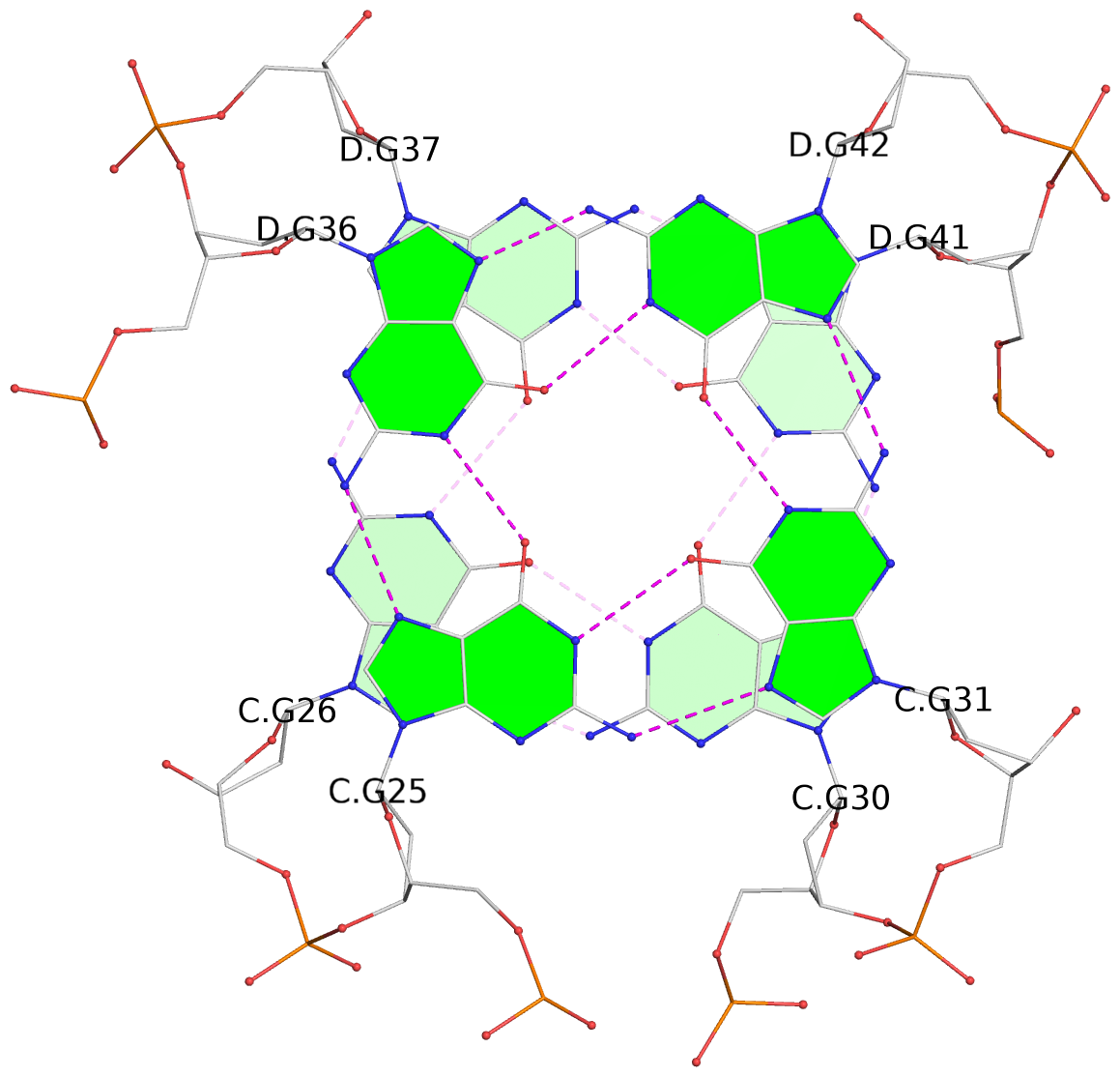
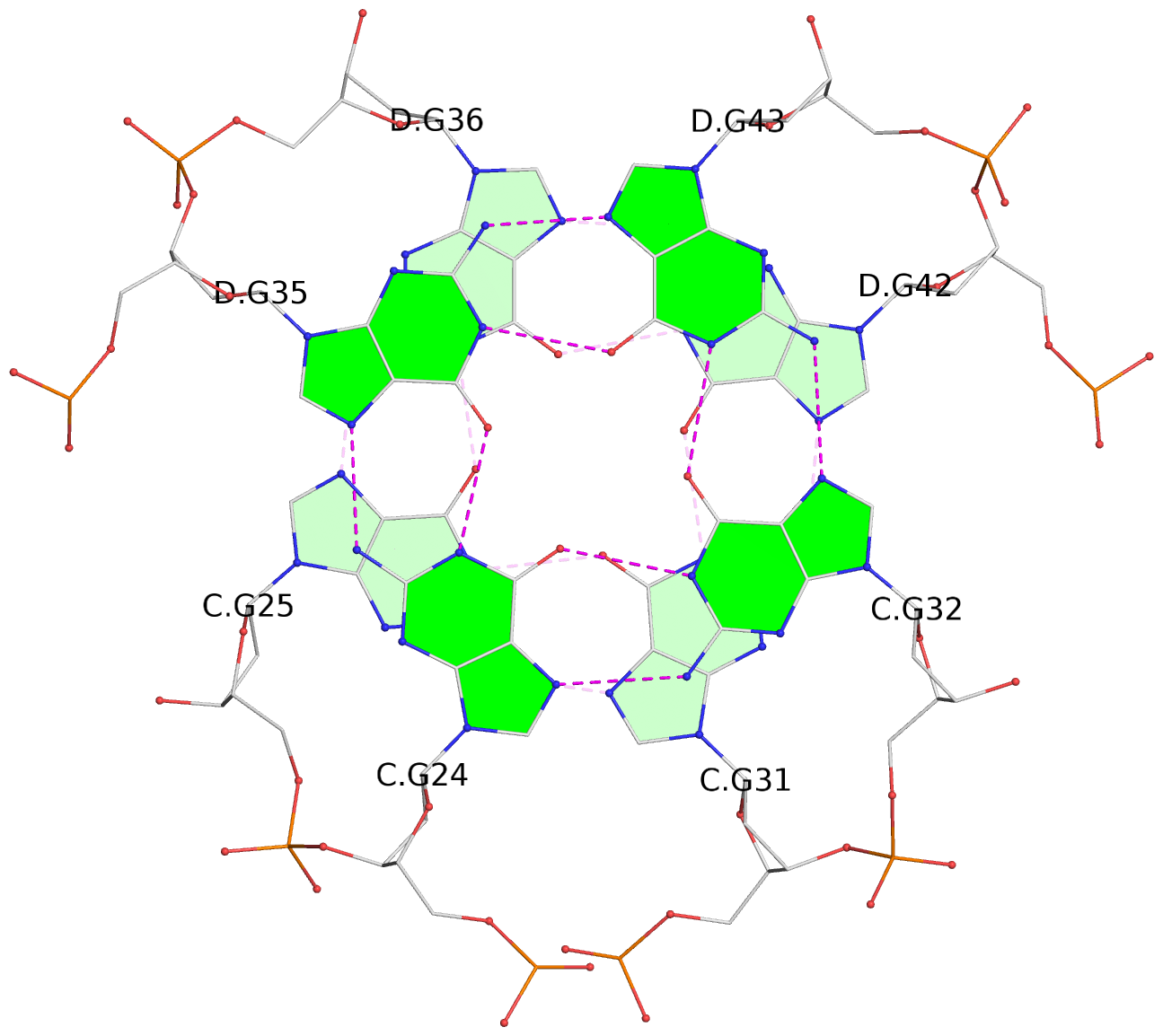
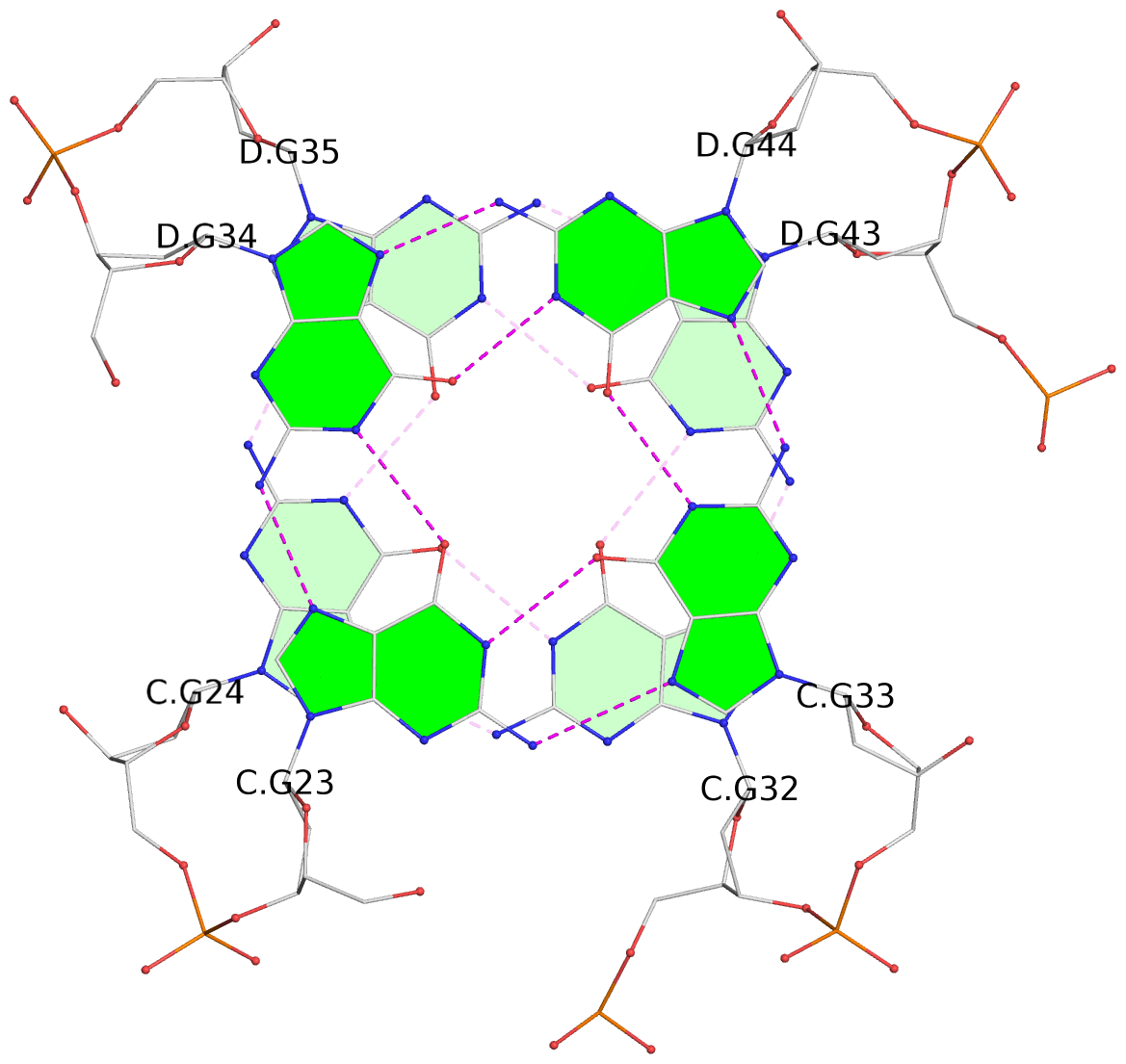
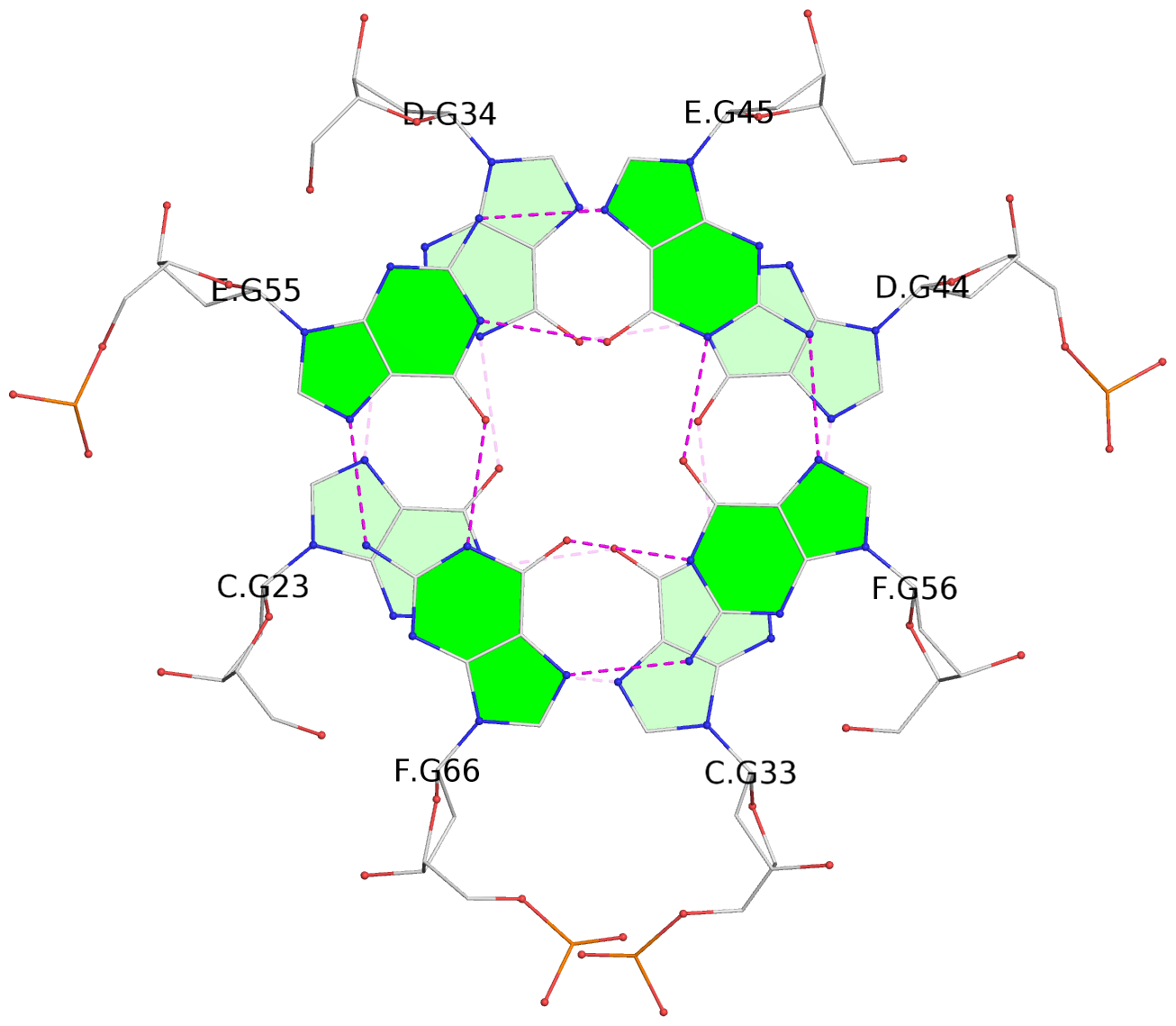
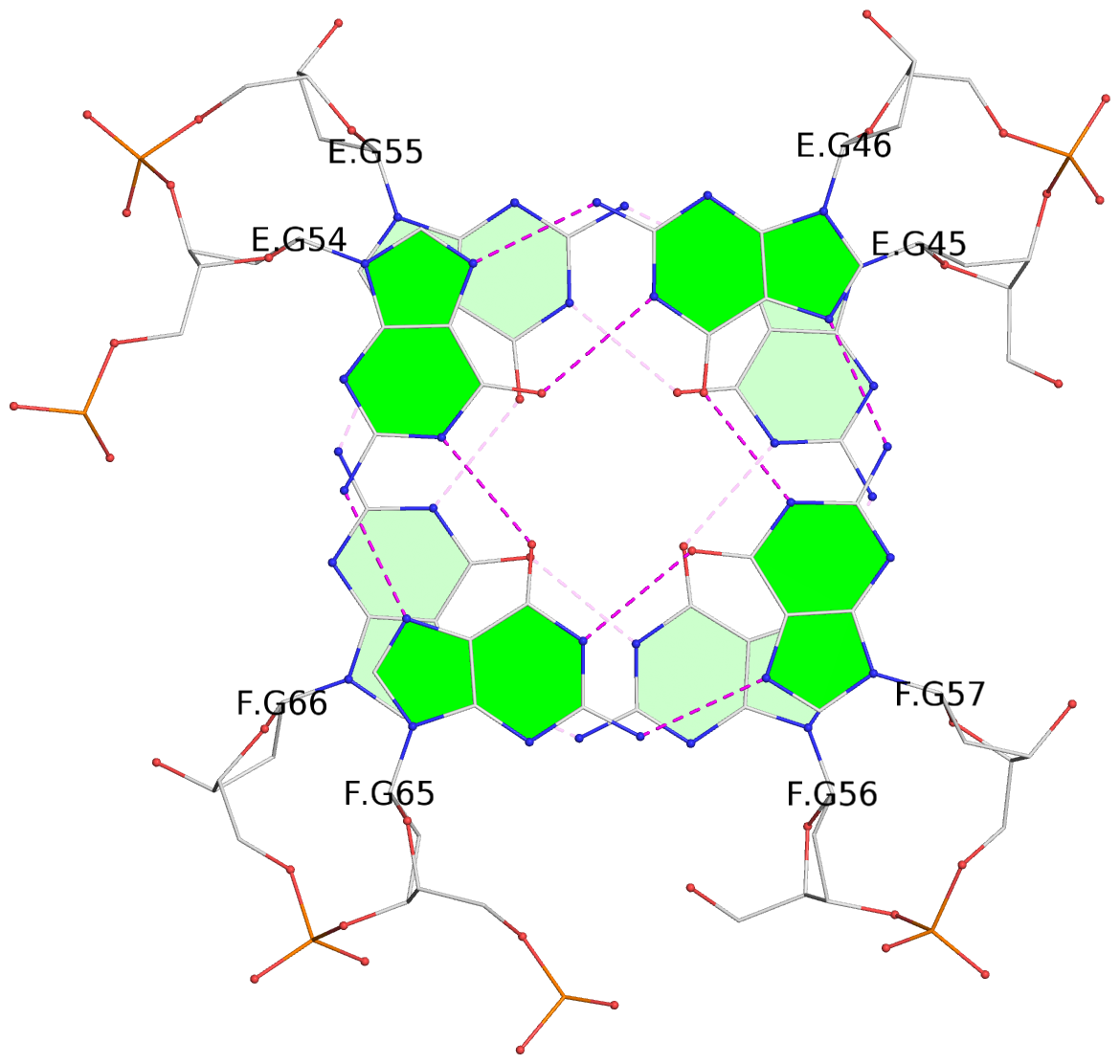
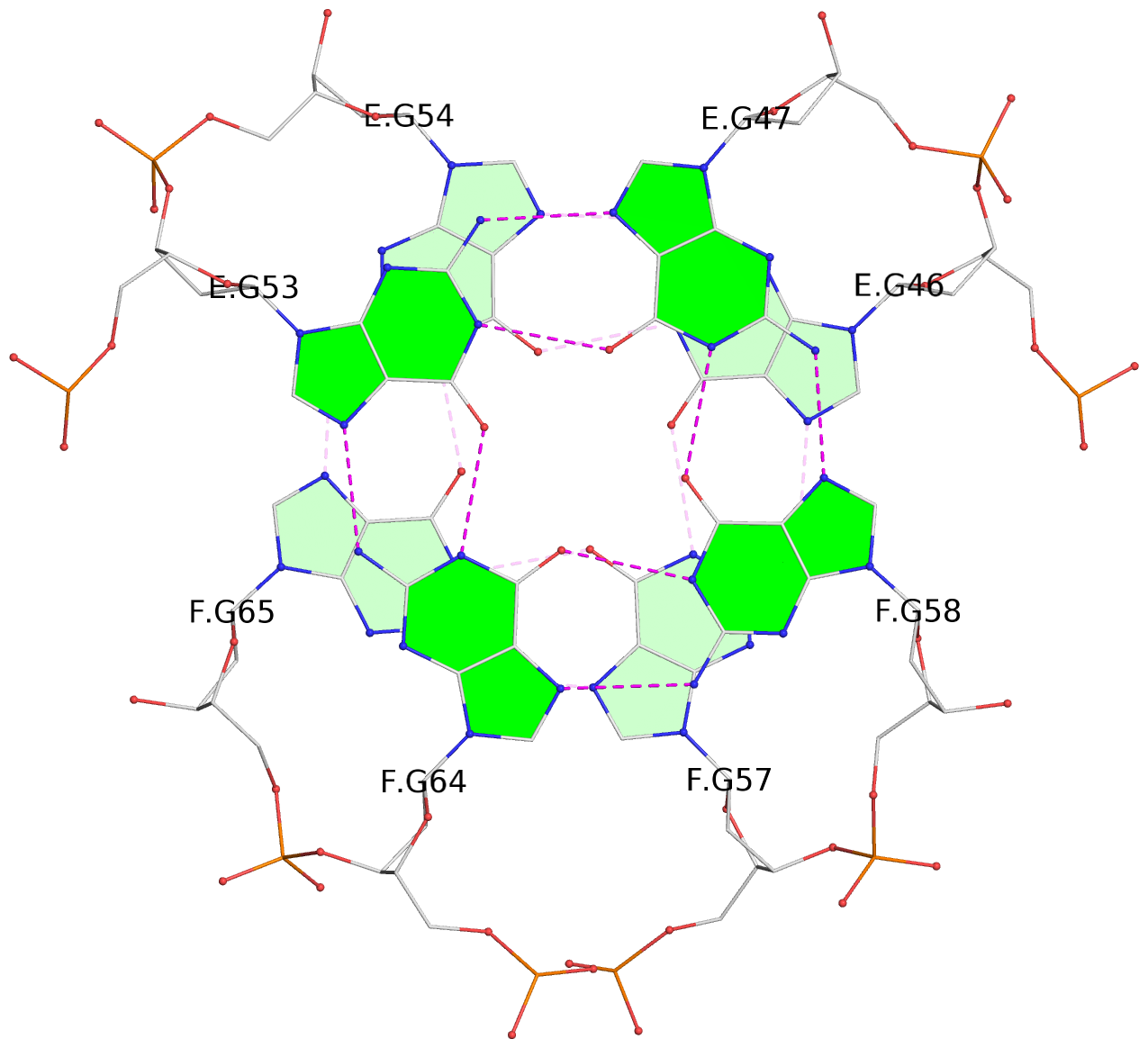
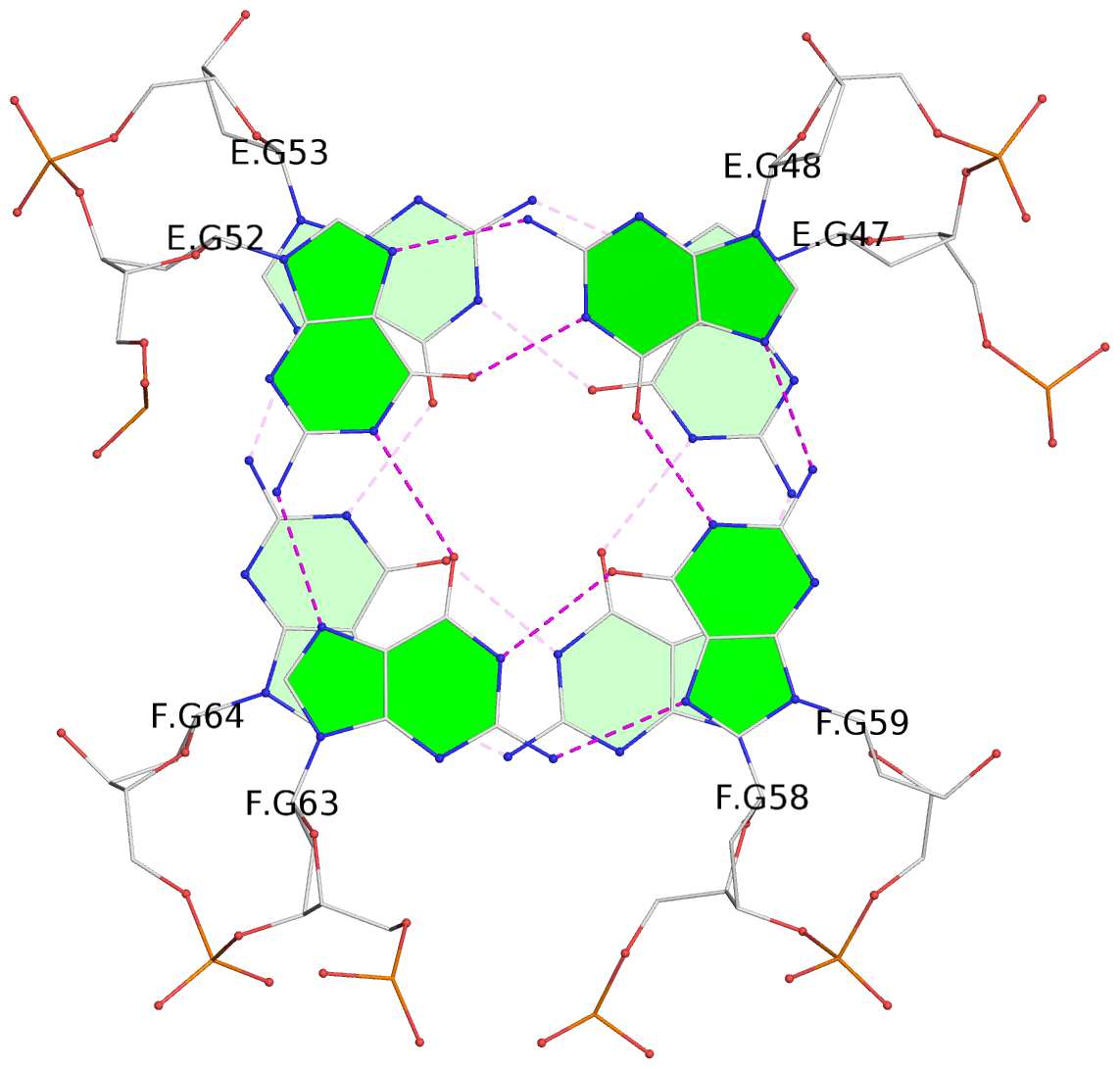
List of 12 G-tetrads
1 glyco-bond=s-s- sugar=---- groove=wnwn planarity=0.452 type=bowl nts=4 GGGG A.DG1,A.DG11,B.DG19,B.DG15 2 glyco-bond=-s-s sugar=---- groove=wnwn planarity=0.343 type=other nts=4 GGGG A.DG2,A.DG10,B.DG20,B.DG14 3 glyco-bond=s-s- sugar=---- groove=wnwn planarity=0.282 type=other nts=4 GGGG A.DG3,A.DG9,B.DG21,B.DG13 4 glyco-bond=-s-s sugar=---- groove=wnwn planarity=0.411 type=bowl nts=4 GGGG A.DG4,A.DG8,B.DG22,B.DG12 5 glyco-bond=ss-- sugar=---- groove=-w-n planarity=0.131 type=planar nts=4 GGGG C.DG23,D.DG34,D.DG44,C.DG33 6 glyco-bond=--ss sugar=---- groove=-w-n planarity=0.094 type=planar nts=4 GGGG C.DG24,D.DG35,D.DG43,C.DG32 7 glyco-bond=ss-- sugar=---- groove=-w-n planarity=0.148 type=planar nts=4 GGGG C.DG25,D.DG36,D.DG42,C.DG31 8 glyco-bond=--ss sugar=---- groove=-w-n planarity=0.245 type=bowl-2 nts=4 GGGG C.DG26,D.DG37,D.DG41,C.DG30 9 glyco-bond=s--s sugar=---- groove=w-n- planarity=0.125 type=planar nts=4 GGGG E.DG45,E.DG55,F.DG66,F.DG56 10 glyco-bond=-ss- sugar=---- groove=w-n- planarity=0.084 type=planar nts=4 GGGG E.DG46,E.DG54,F.DG65,F.DG57 11 glyco-bond=s--s sugar=---- groove=w-n- planarity=0.120 type=planar nts=4 GGGG E.DG47,E.DG53,F.DG64,F.DG58 12 glyco-bond=-ss- sugar=---- groove=w-n- planarity=0.258 type=bowl nts=4 GGGG E.DG48,E.DG52,F.DG63,F.DG59
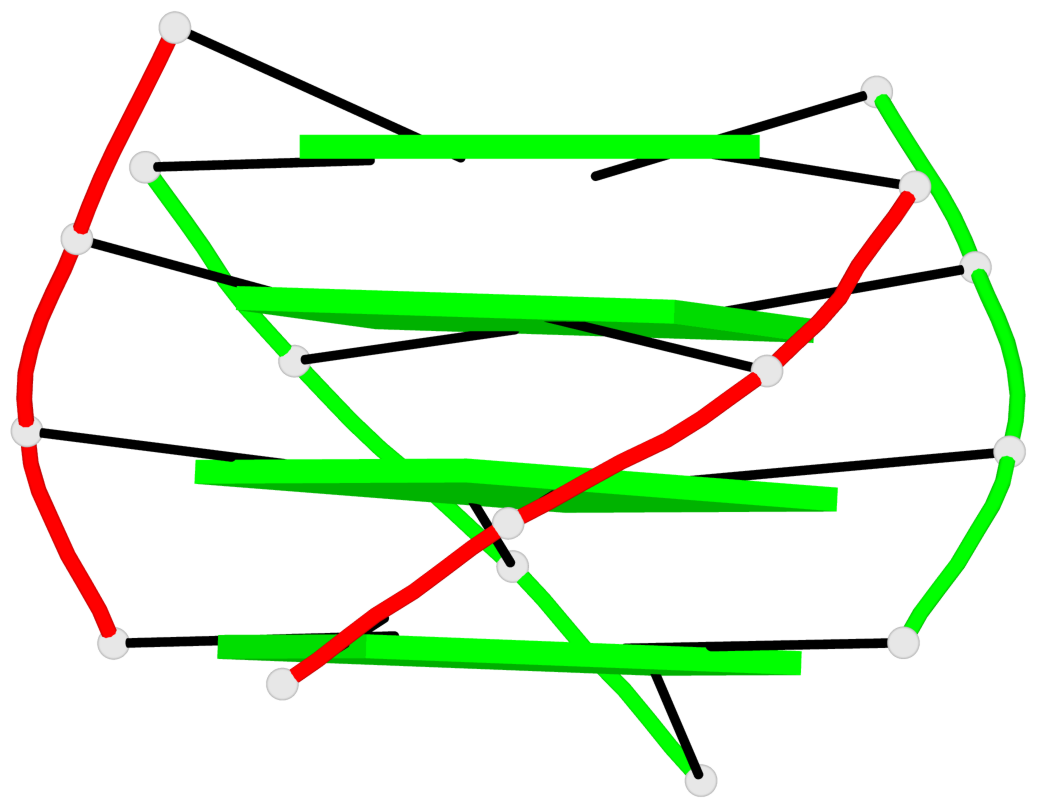
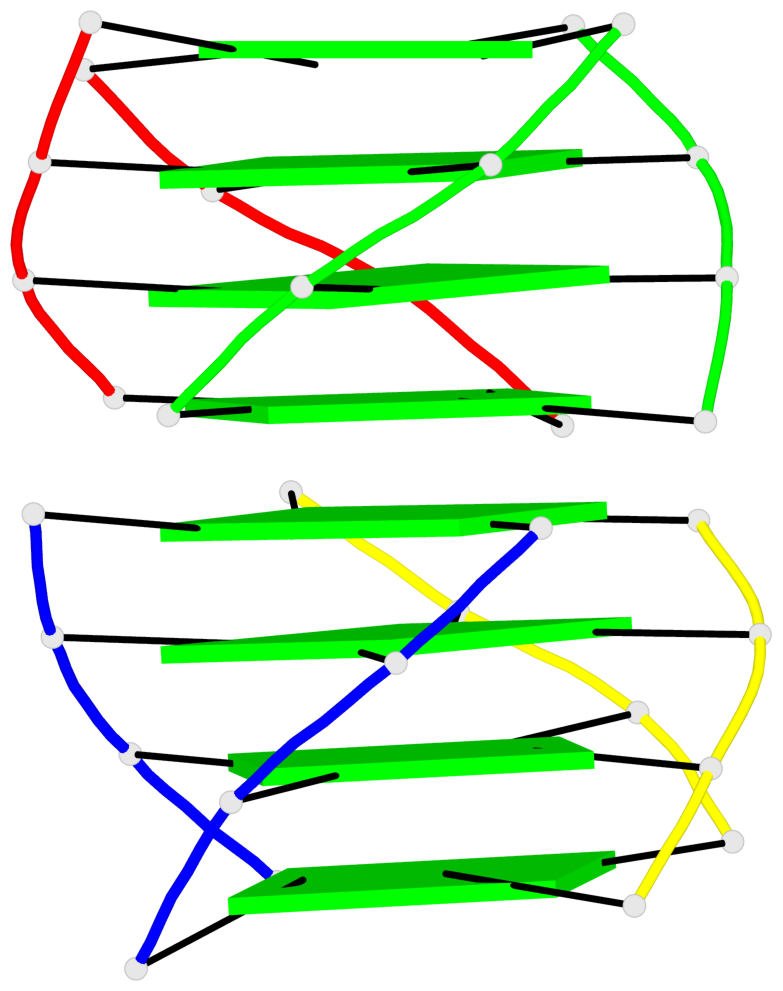
List of 2 G4-helices
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 4 G-tetrad layers, inter-molecular, with 1 stem
Helix#2, 8 G-tetrad layers, inter-molecular, with 2 stems
List of 3 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 4 G-tetrad layers, 2 loops, inter-molecular, UDUD, anti-parallel, (2+2)
Stem#2, 4 G-tetrad layers, 2 loops, inter-molecular, UUDD, anti-parallel, (2+2)
Stem#3, 4 G-tetrad layers, 2 loops, inter-molecular, UDDU, anti-parallel, (2+2)
List of 1 G4 coaxial stack
1 G4 helix#2 contains 2 G4 stems: [#2,#3] [mixed]