Detailed DSSR results for the G-quadruplex: PDB entry 2chk
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 2chk
- Class
- nucleic acid
- Method
- NMR
- Summary
- NMR structure of tllllt quadruplex
- Reference
- Nielsen JT, Arar K, Petersen M (2006): "NMR Solution Structures of Lna (Locked Nucleic Acid) Modified Quadruplexes." Nucleic Acids Res., 34, 2006. doi: 10.1093/NAR/GKL144.
- Abstract
- We have determined the NMR solution structures of the quadruplexes formed by d(TGLGLT) and d(TL4T), where L denotes LNA (locked nucleic acid) modified G-residues. Both structures are tetrameric, parallel and right-handed and the native global fold of the corresponding DNA quadruplex is retained upon introduction of the LNA nucleotides. However, local structural alterations are observed owing to the locked LNA sugars. In particular, a distinct change in the sugar-phosphate backbone is observed at the G2pL3 and L2pL3 base steps and sequence dependent changes in the twist between tetrads are also seen. Both the LNA modified quadruplexes have raised thermostability as compared to the DNA quadruplex. The quadruplex-forming capability of d(TGLGLT) is of particular interest as it expands the design flexibility for stable parallel LNA quadruplexes and shows that LNA nucleotides can be mixed with DNA or other modified nucleic acids. As such, LNA-based quadruplexes can be decorated by a variety of chemical modifications. Such LNA quadruplex scaffolds might find applications in the developing field of nanobiotechnology.
- G4 notes
- 4 G-tetrads, 1 G4 helix, 1 G4 stem, parallel(4+0), UUUU
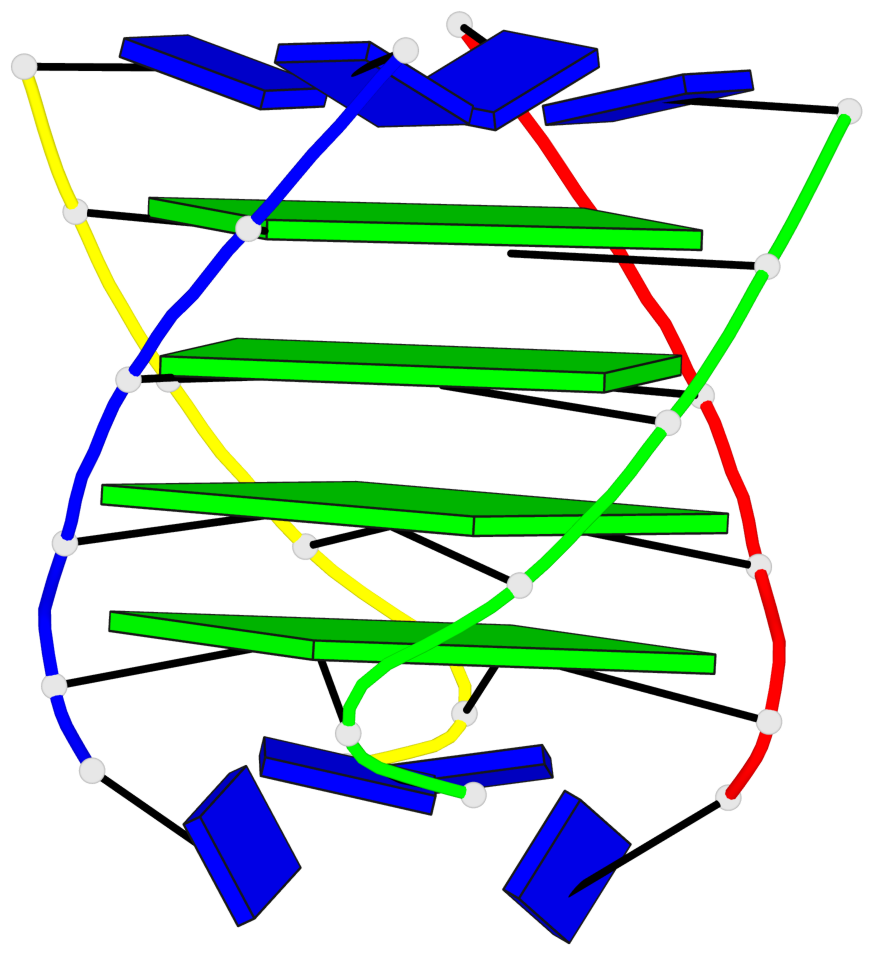
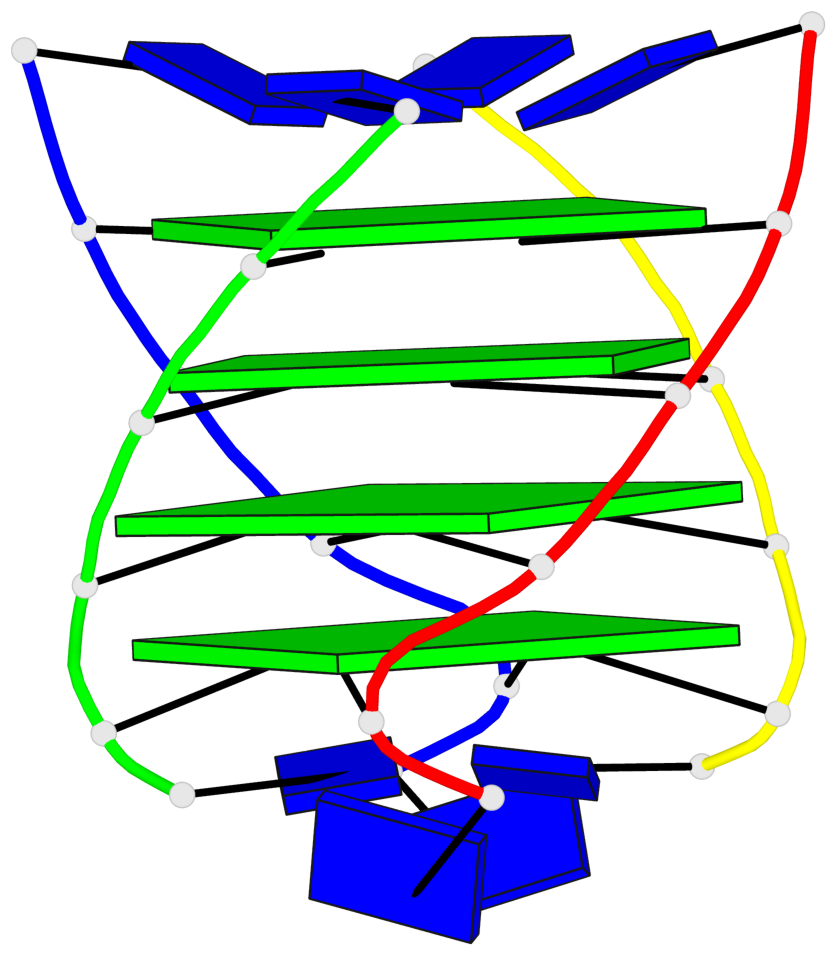
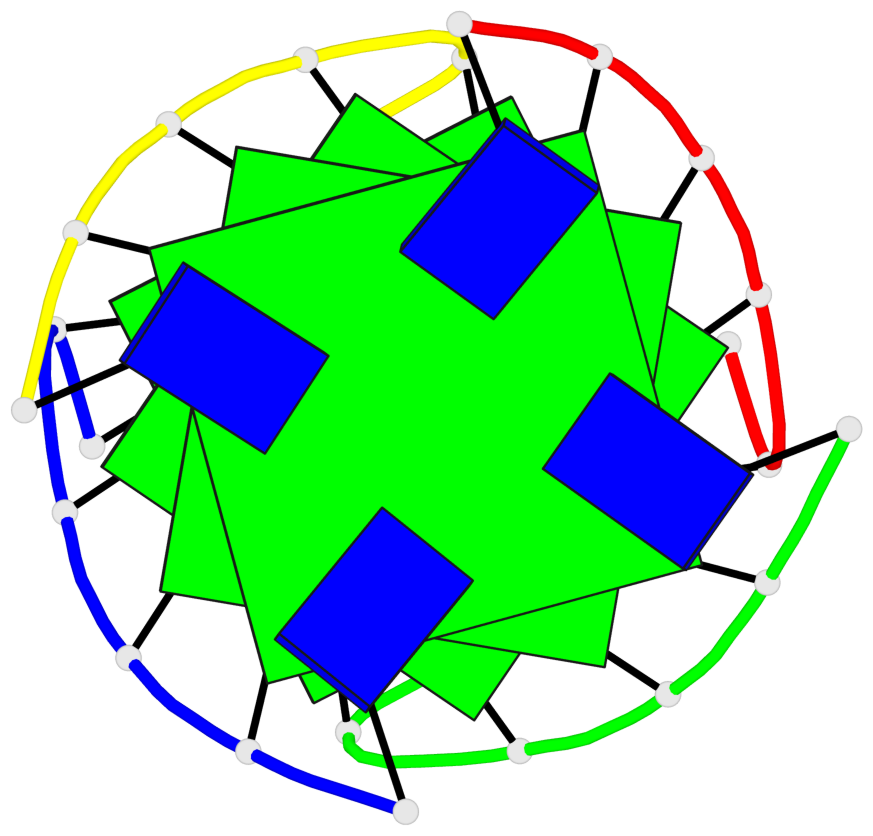
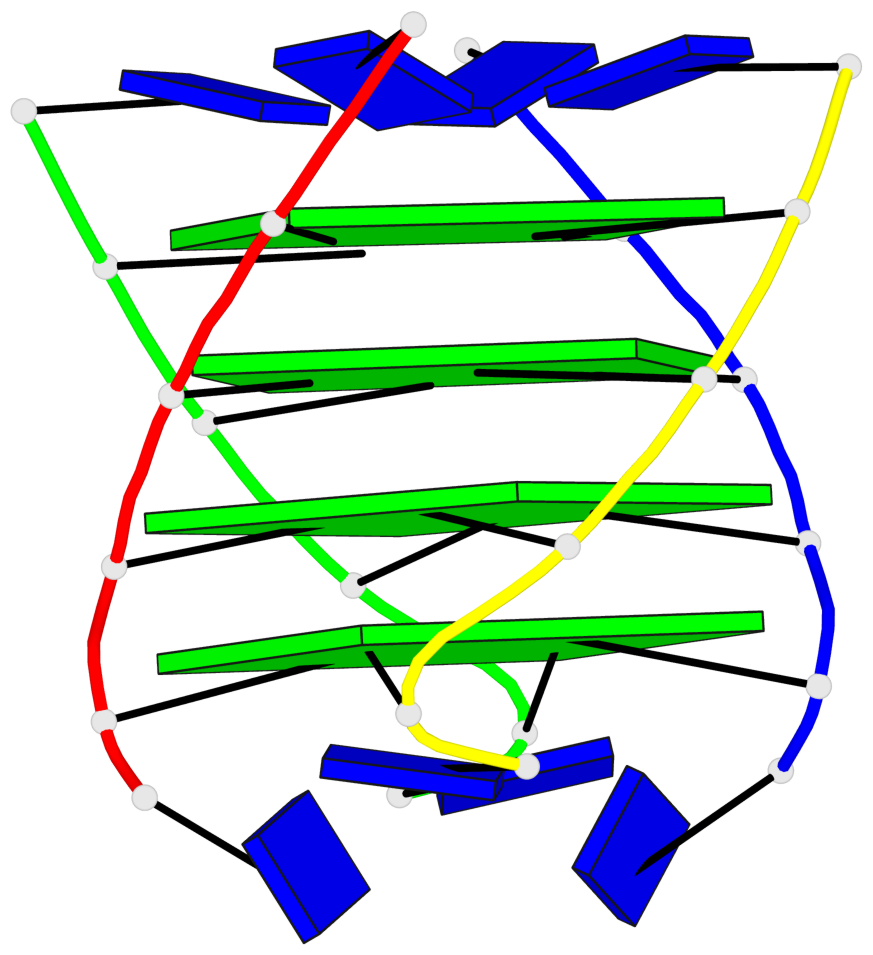
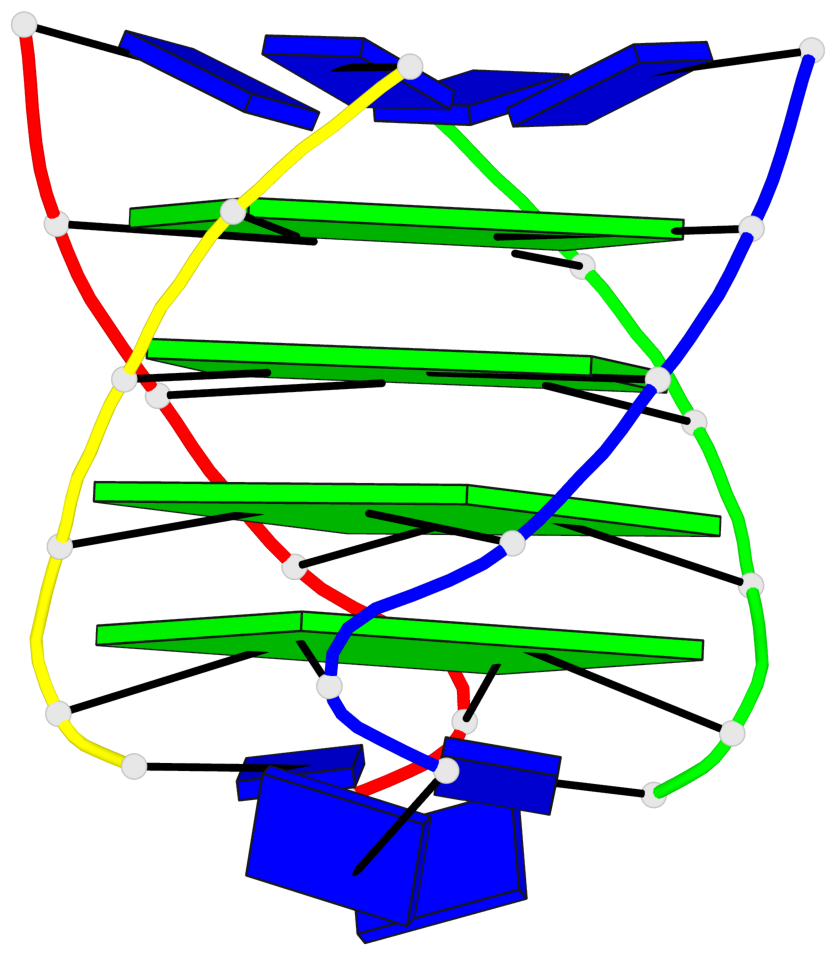
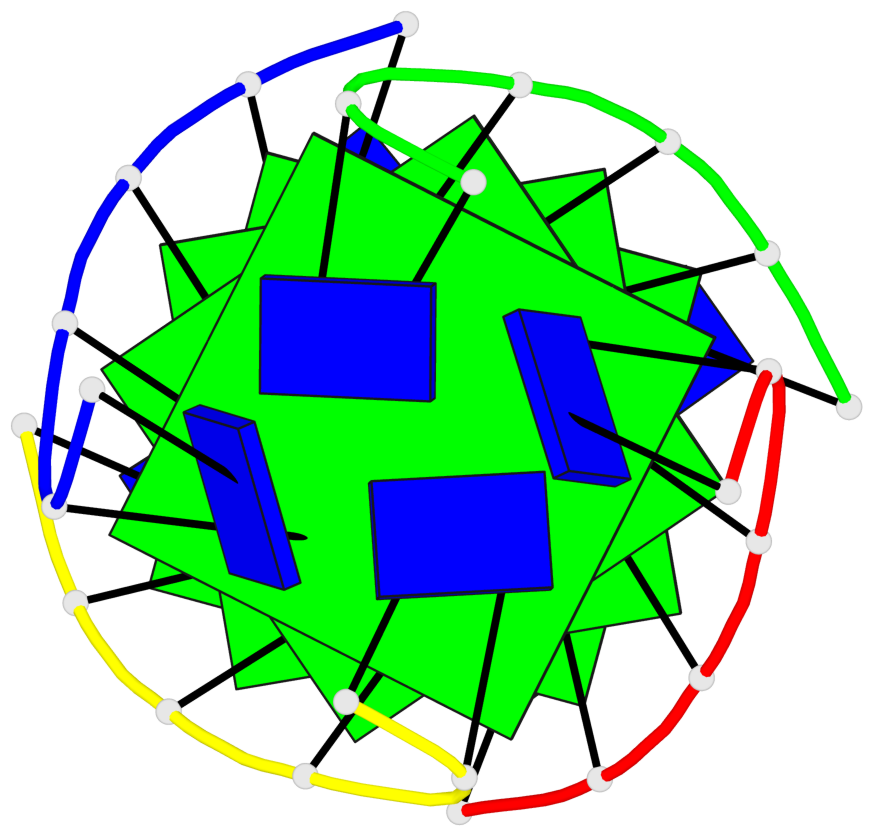
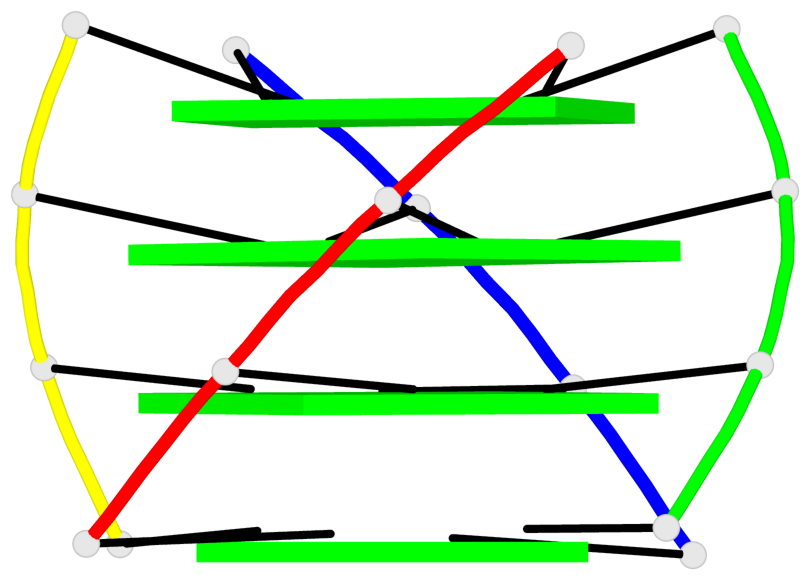
Base-block schematics in six views
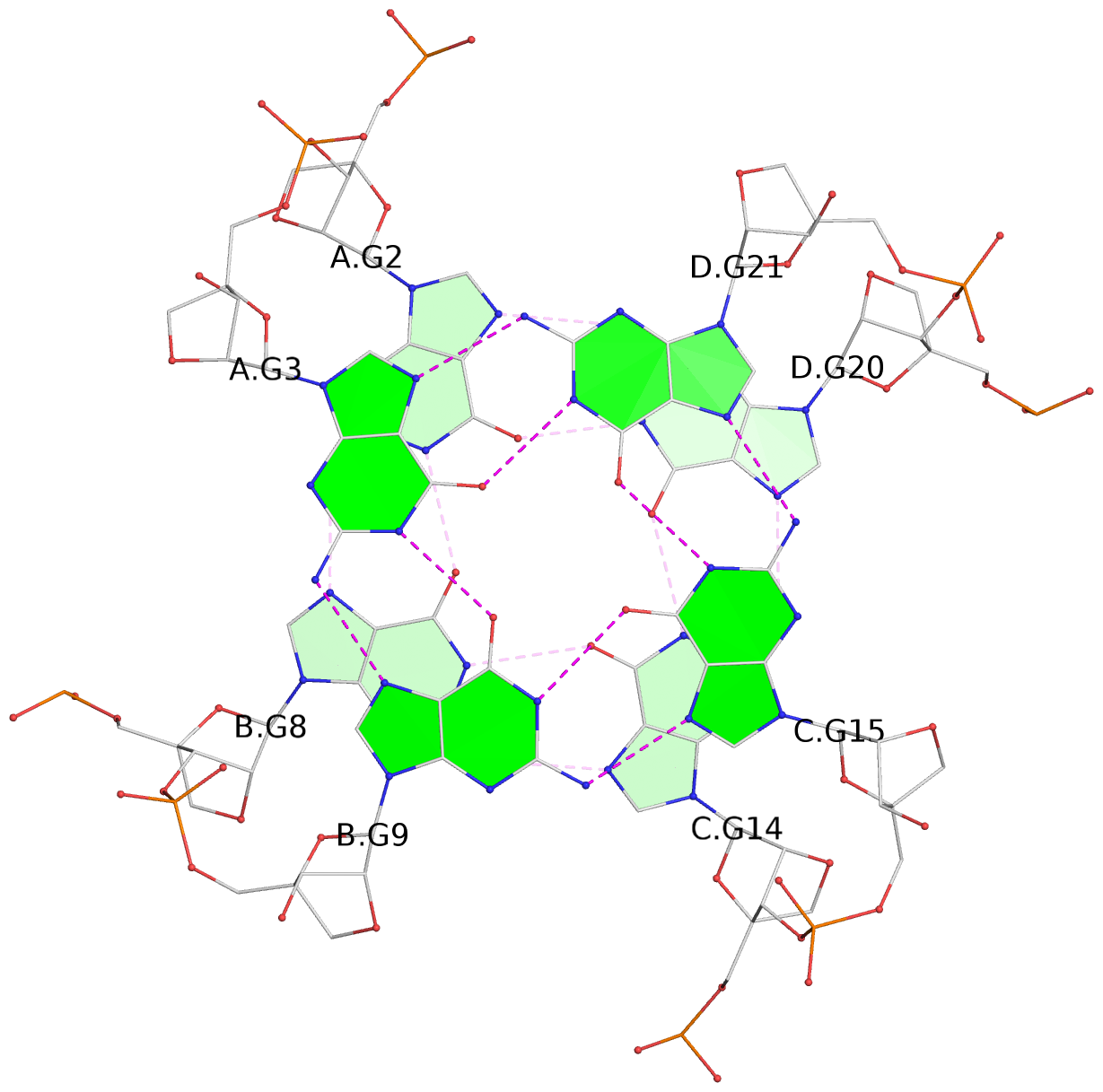
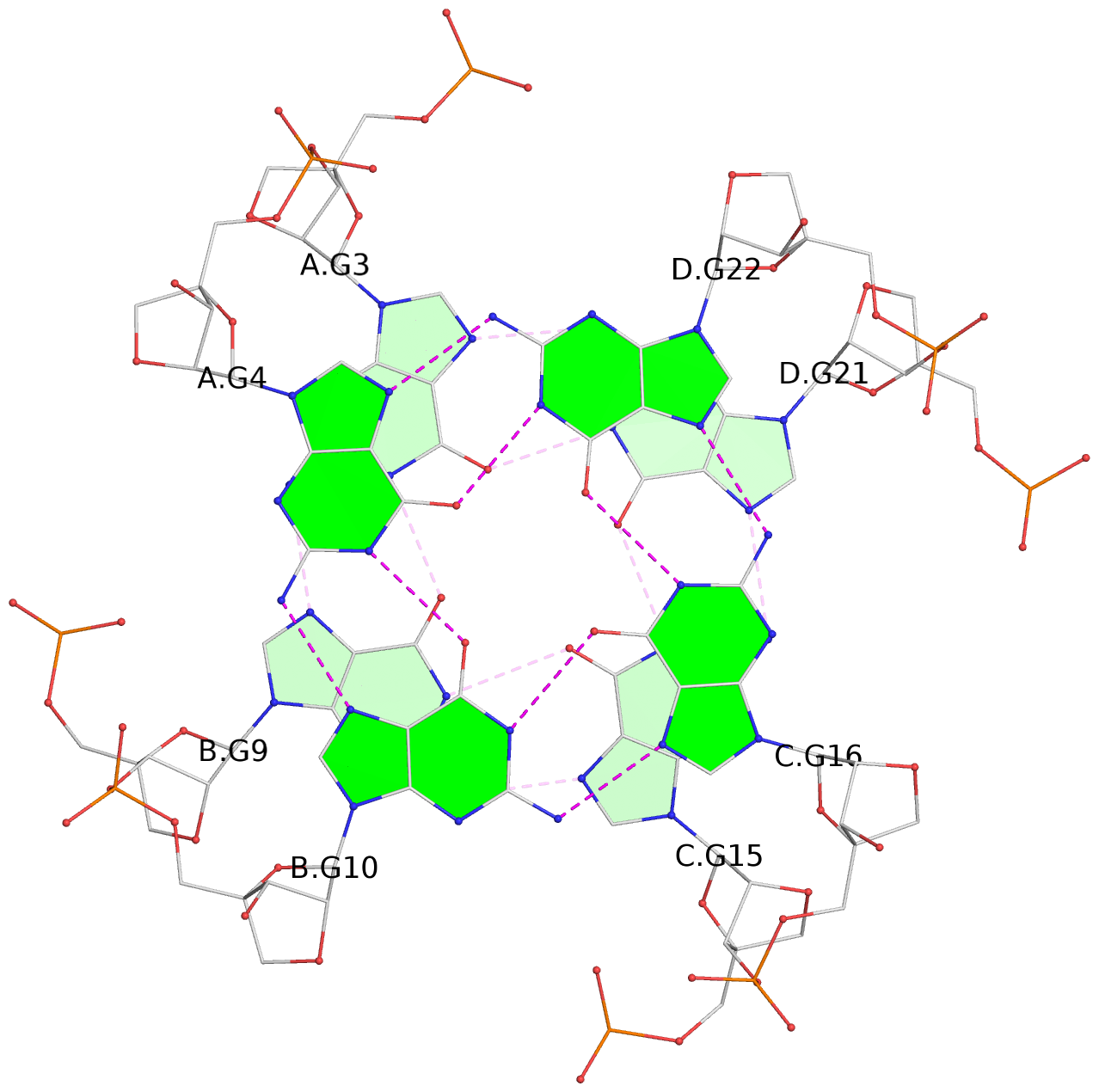
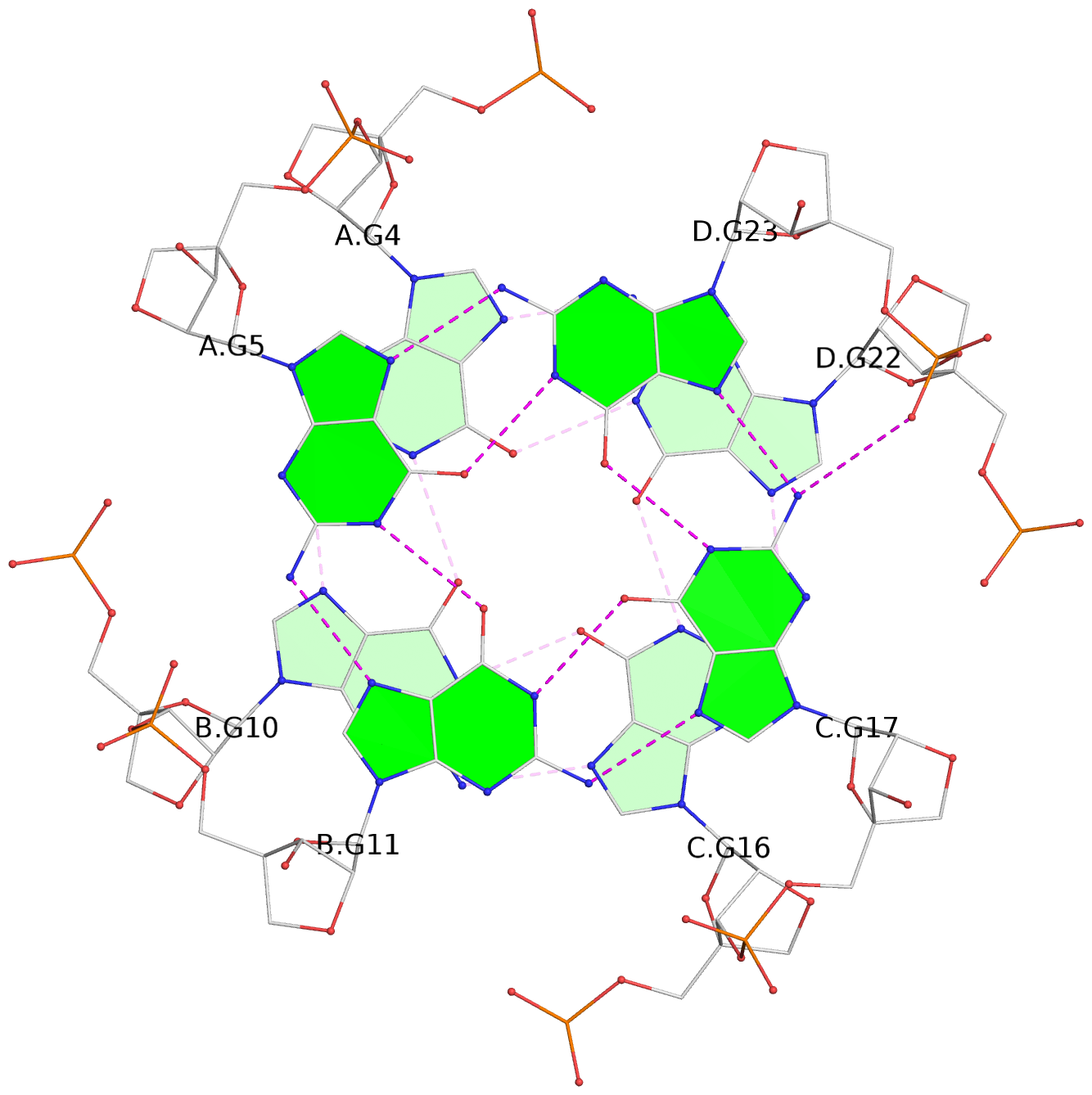
List of 4 G-tetrads
1 glyco-bond=---- sugar=3333 groove=---- planarity=0.514 type=other nts=4 gggg A.LCG2,B.LCG8,C.LCG14,D.LCG20 2 glyco-bond=---- sugar=3333 groove=---- planarity=0.350 type=other nts=4 gggg A.LCG3,B.LCG9,C.LCG15,D.LCG21 3 glyco-bond=---- sugar=3333 groove=---- planarity=0.212 type=other nts=4 gggg A.LCG4,B.LCG10,C.LCG16,D.LCG22 4 glyco-bond=---- sugar=3333 groove=---- planarity=0.164 type=other nts=4 gggg A.LCG5,B.LCG11,C.LCG17,D.LCG23
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 4 G-tetrad layers, inter-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.