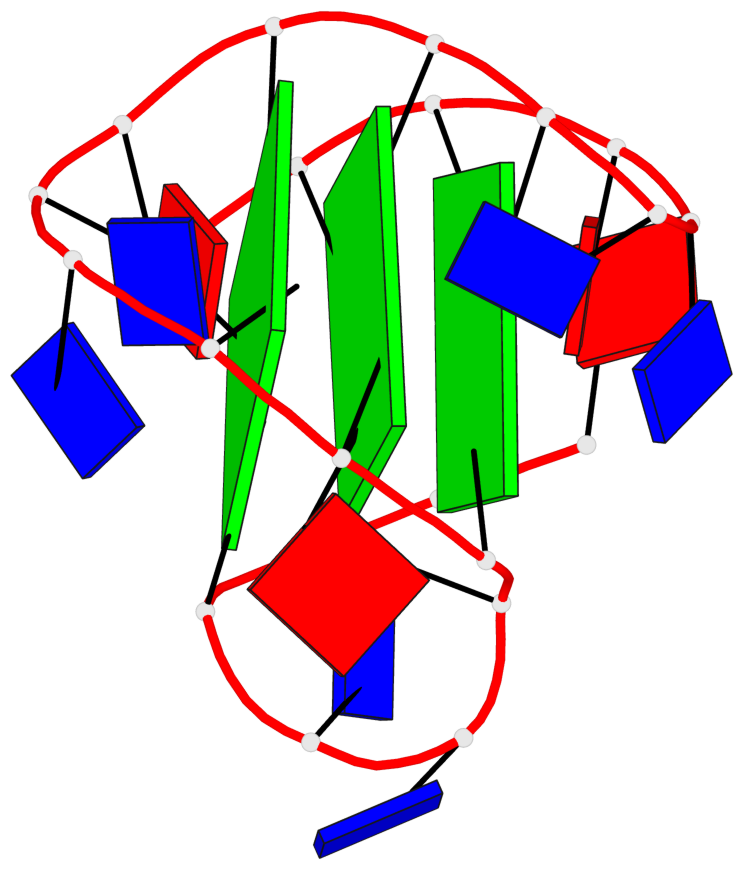
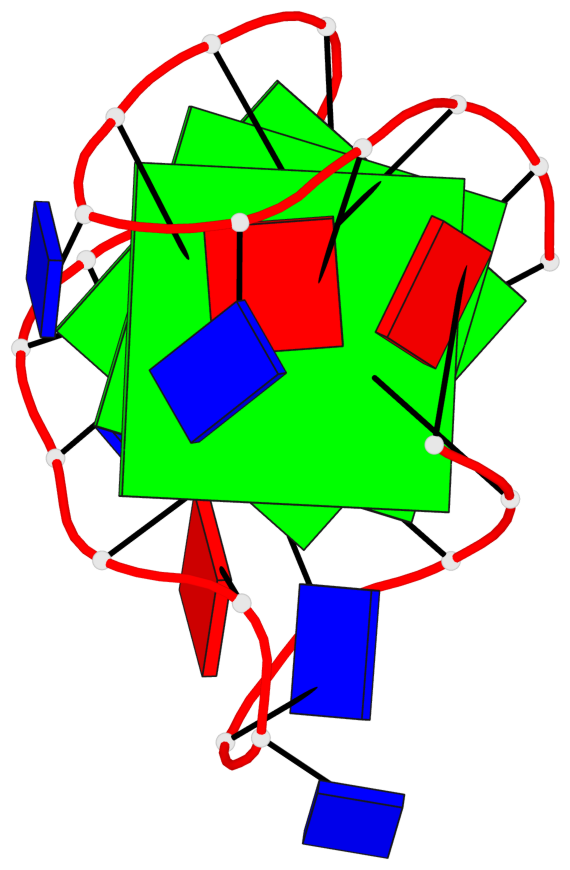
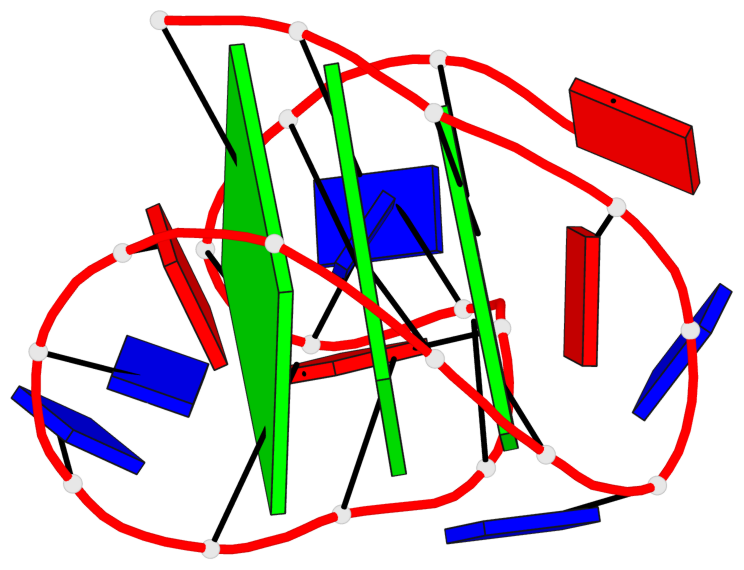
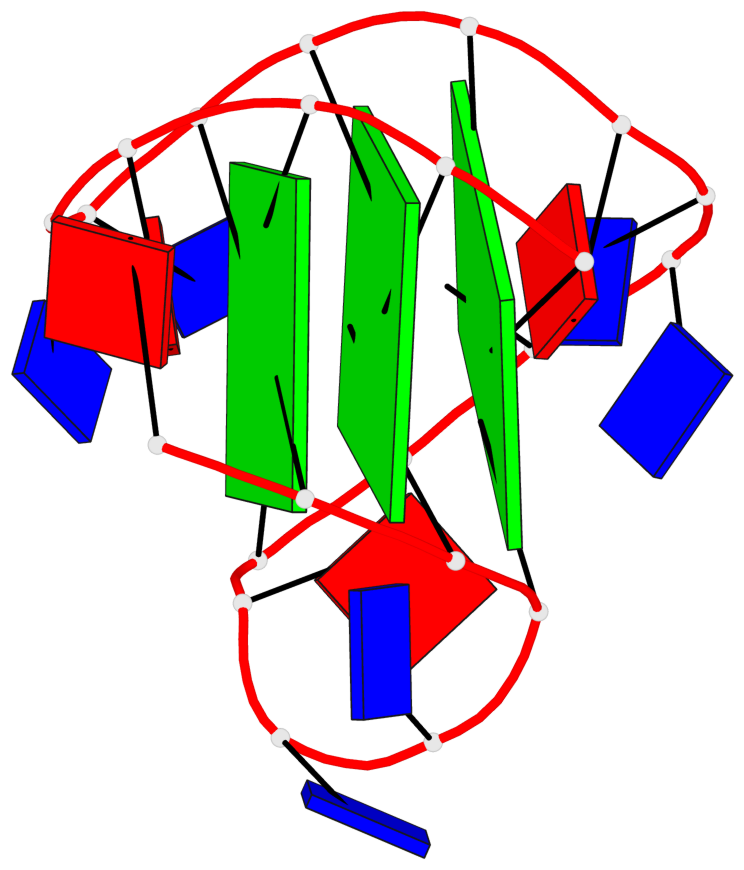
Detailed DSSR results for the G-quadruplex: PDB entry 2e4i
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 2e4i
- Class
- DNA
- Method
- NMR
- Summary
- Human telomeric DNA mixed-parallel-antiparallel quadruplex under physiological ionic conditions stabilized by proper incorporation of 8-bromoguanosines
- Reference
- Matsugami A, Xu Y, Noguchi Y, Sugiyama H, Katahira M (2007): "Structure of a human telomeric DNA sequence stabilized by 8-bromoguanosine substitutions, as determined by NMR in a K+ solution." Febs J., 274, 3545-3556. doi: 10.1111/j.1742-4658.2007.05881.x.
- Abstract
- The structure of human telomeric DNA is controversial; it depends upon the sequence contexts and the methodologies used to determine it. The solution structure in the presence of K(+) is particularly interesting, but the structure is yet to be elucidated, due to possible conformational heterogeneity. Here, a unique strategy is applied to stabilize one such structure in a K(+) solution by substituting guanosines with 8-bromoguanosines at proper positions. The resulting spectra are cleaner and led to determination of the structure at a high atomic resolution. This demonstrates that the application of 8-bromoguanosine is a powerful tool to overcome the difficulty of nucleic acid structure determination arising from conformational heterogeneity. The obtained structure is a mixed-parallel/antiparallel quadruplex. The structure of telomeric DNA was recently reported in another study, in which stabilization was brought about by mutation and resultant additional interactions [Luu KN, Phan AT, Kuryavyi V, Lacroix L & Patel DJ (2006) Structure of the human telomere in K(+) solution: an intramolecular (3+1) G-quadruplex scaffold. J Am Chem Soc 128, 9963-9970]. The structure of the guanine tracts was similar between the two. However, a difference was seen for loops connecting guanine tracts, which may play a role in the higher order arrangement of telomeres. Our structure can be utilized to design a small molecule which stabilizes the quadruplex. This type of molecule is supposed to inhibit a telomerase and thus is expected to be a candidate anticancer drug.
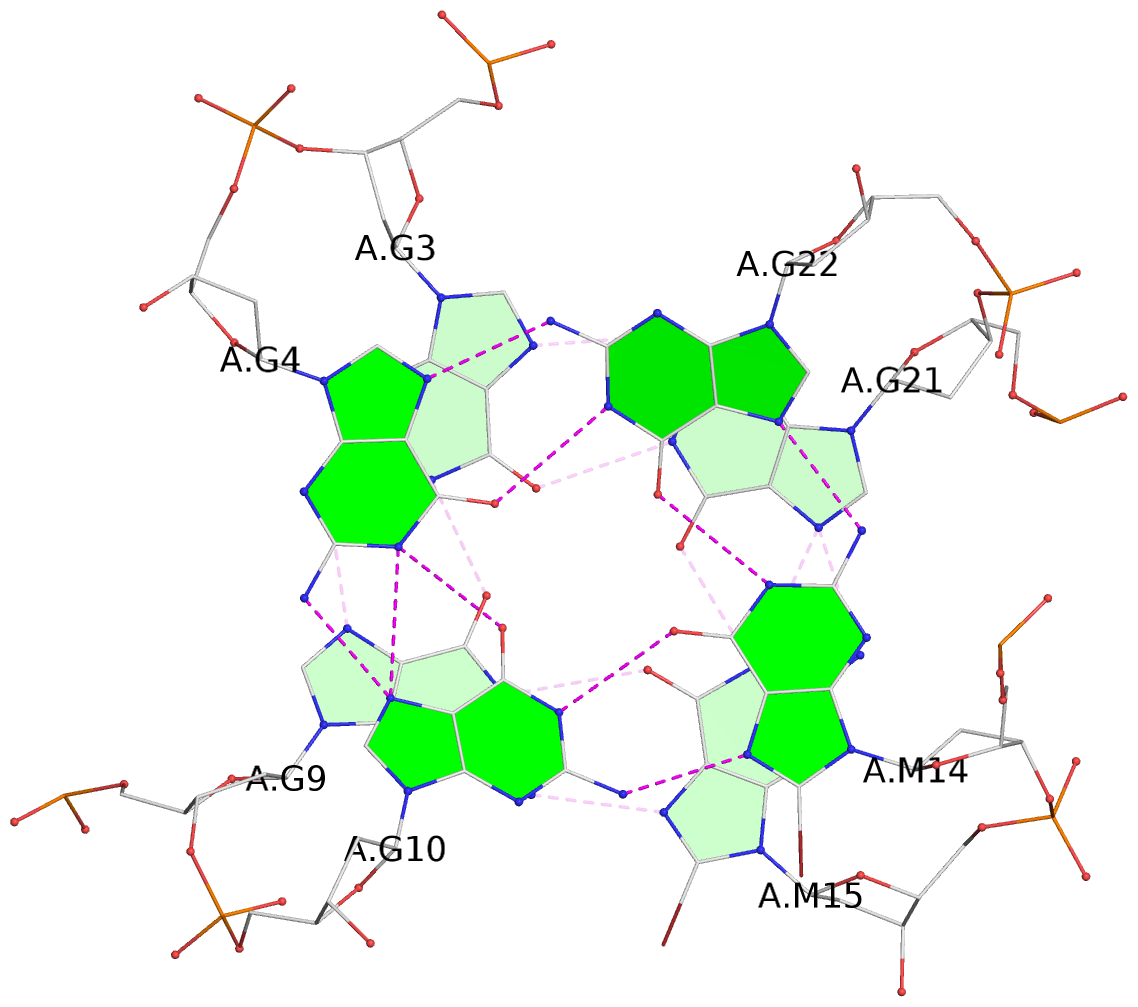
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(-P-Lw-Ln), hybrid-1(3+1), UUDU
Base-block schematics in six views
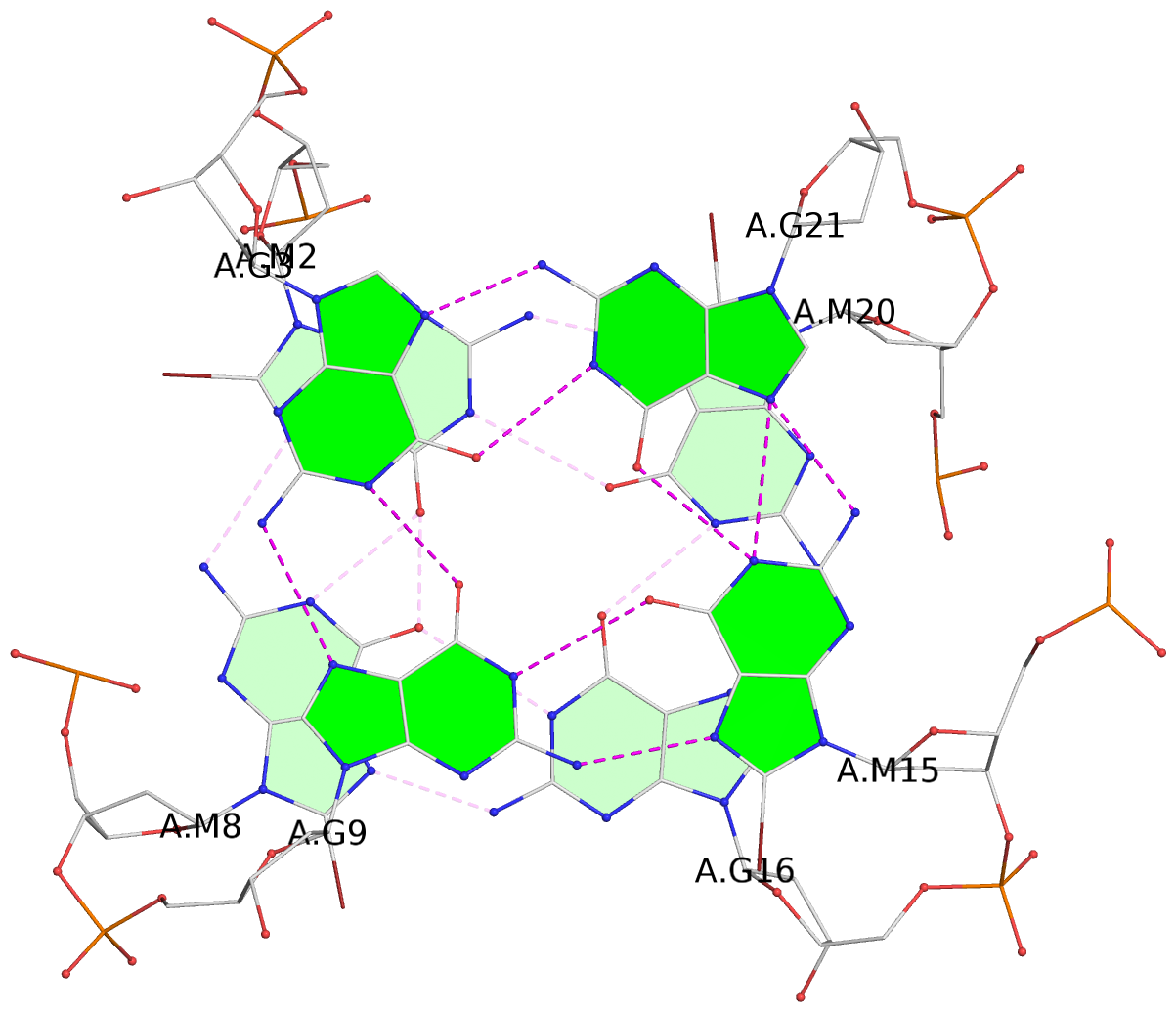
List of 3 G-tetrads
1 glyco-bond=ss-s sugar=---- groove=-wn- planarity=0.238 type=other nts=4 ggGg A.BGM2,A.BGM8,A.DG16,A.BGM20 2 glyco-bond=--s- sugar=-.-- groove=-wn- planarity=0.336 type=other nts=4 GGgG A.DG3,A.DG9,A.BGM15,A.DG21 3 glyco-bond=--s- sugar=---. groove=-wn- planarity=0.459 type=bowl nts=4 GGgG A.DG4,A.DG10,A.BGM14,A.DG22
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.