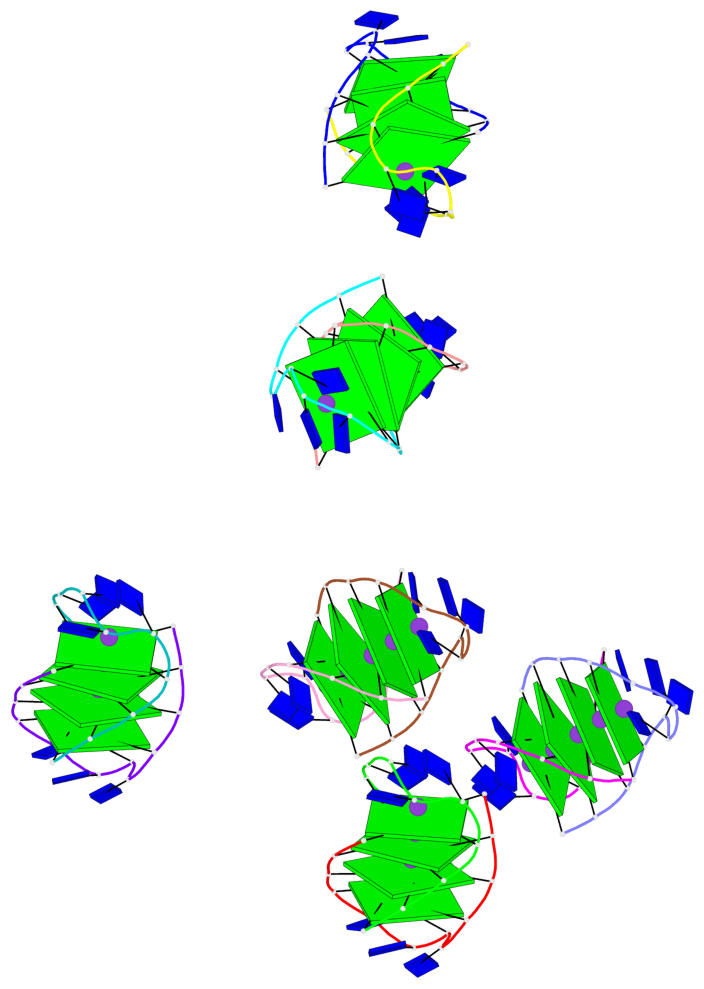
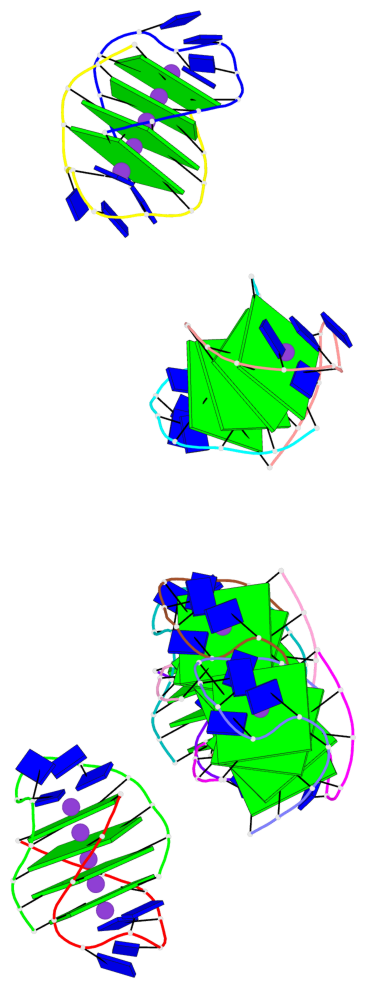
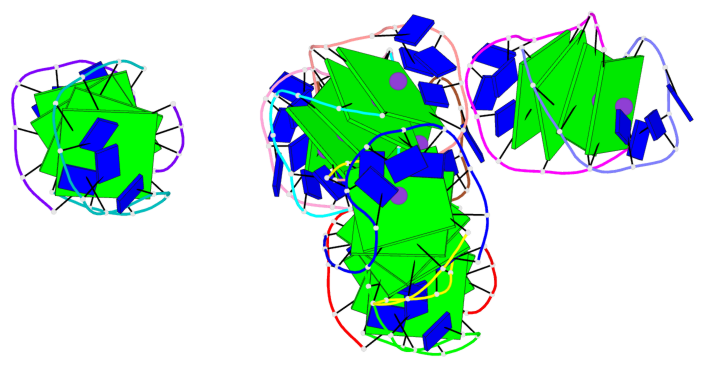
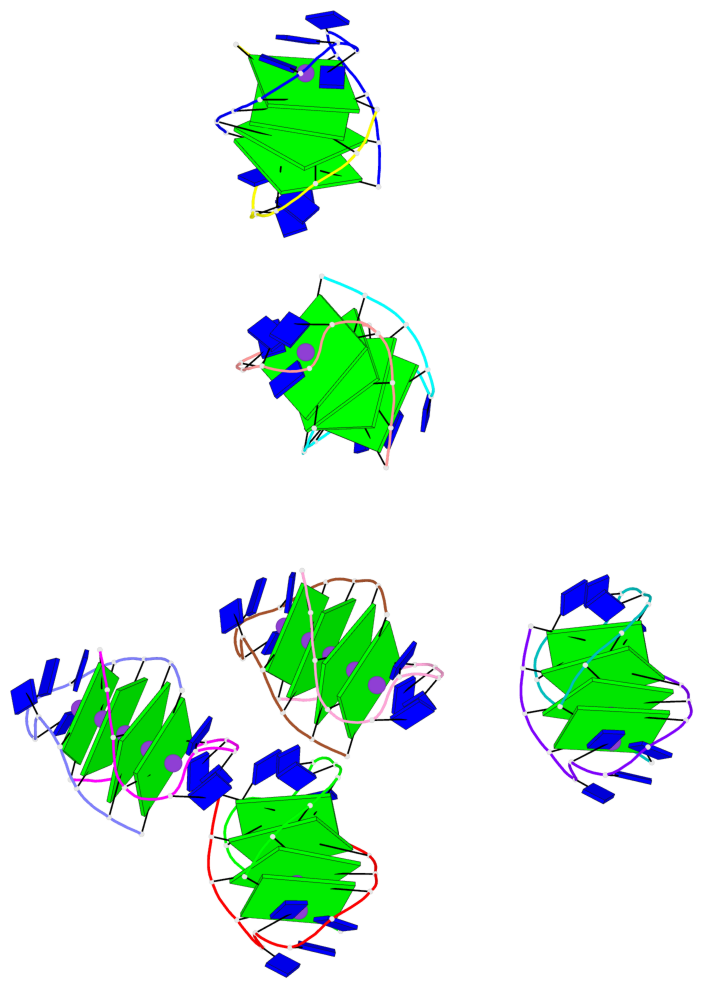
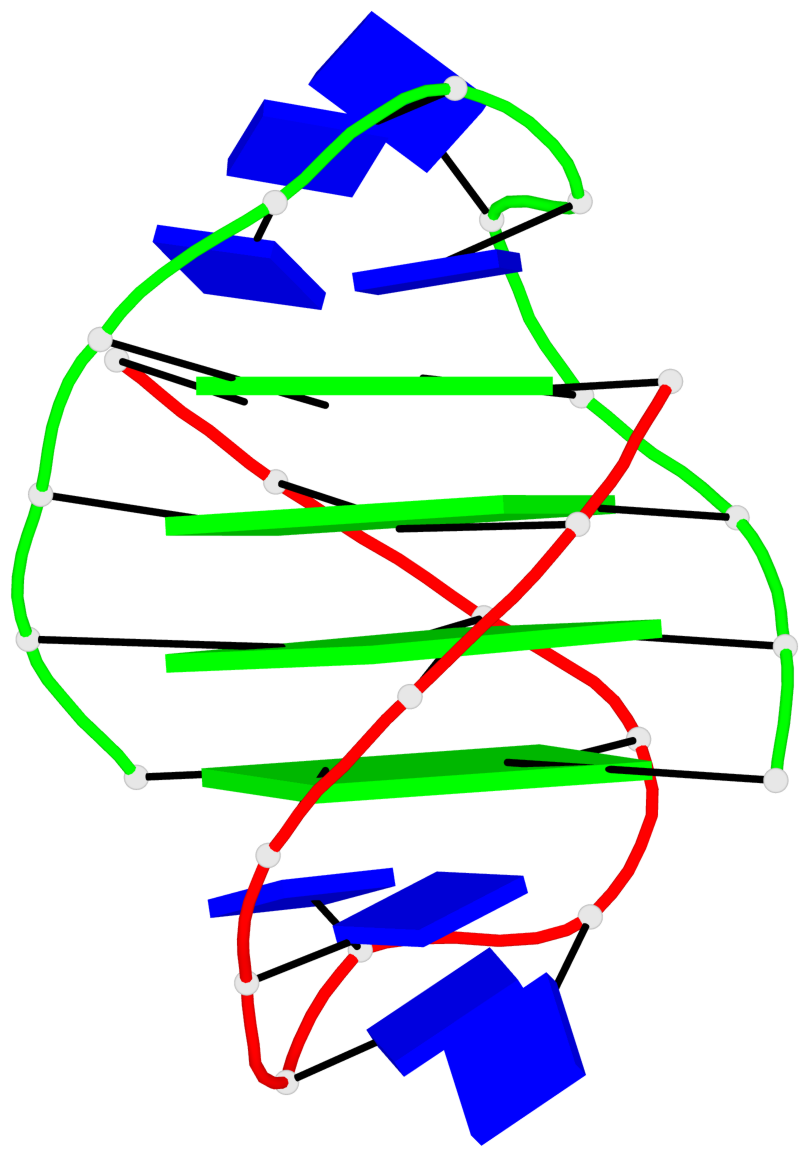
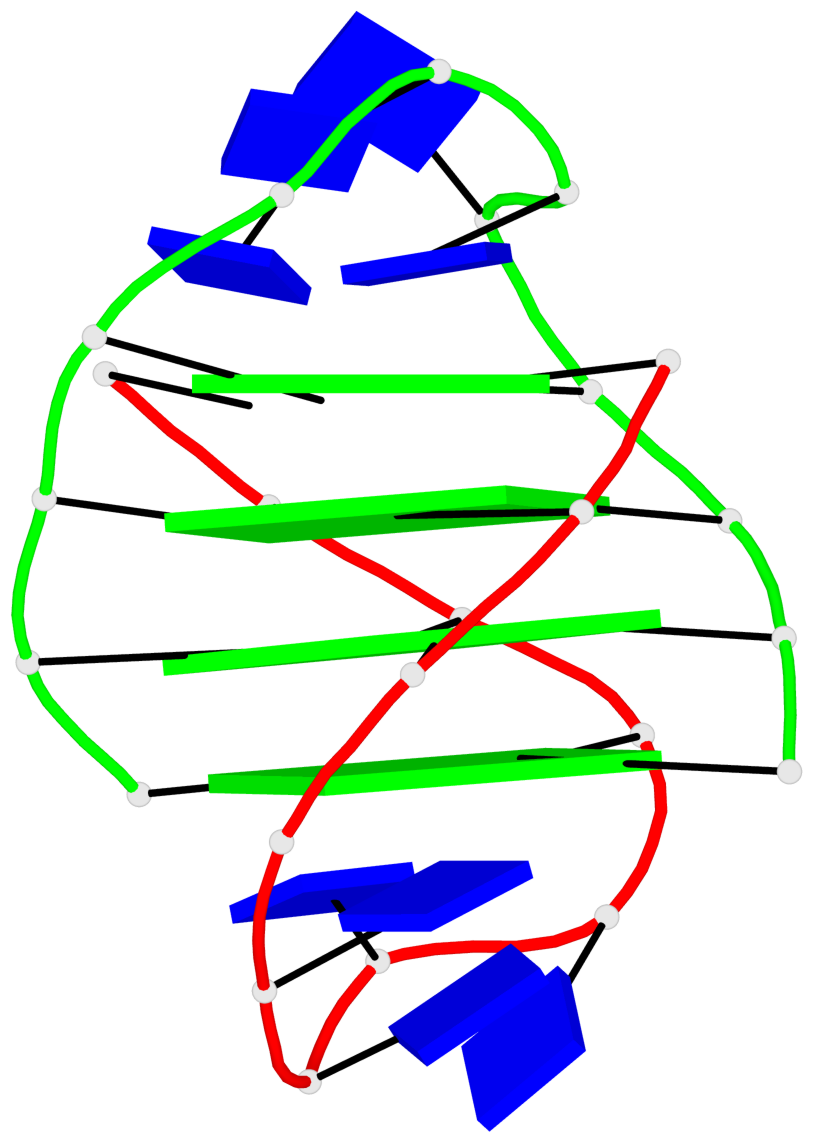
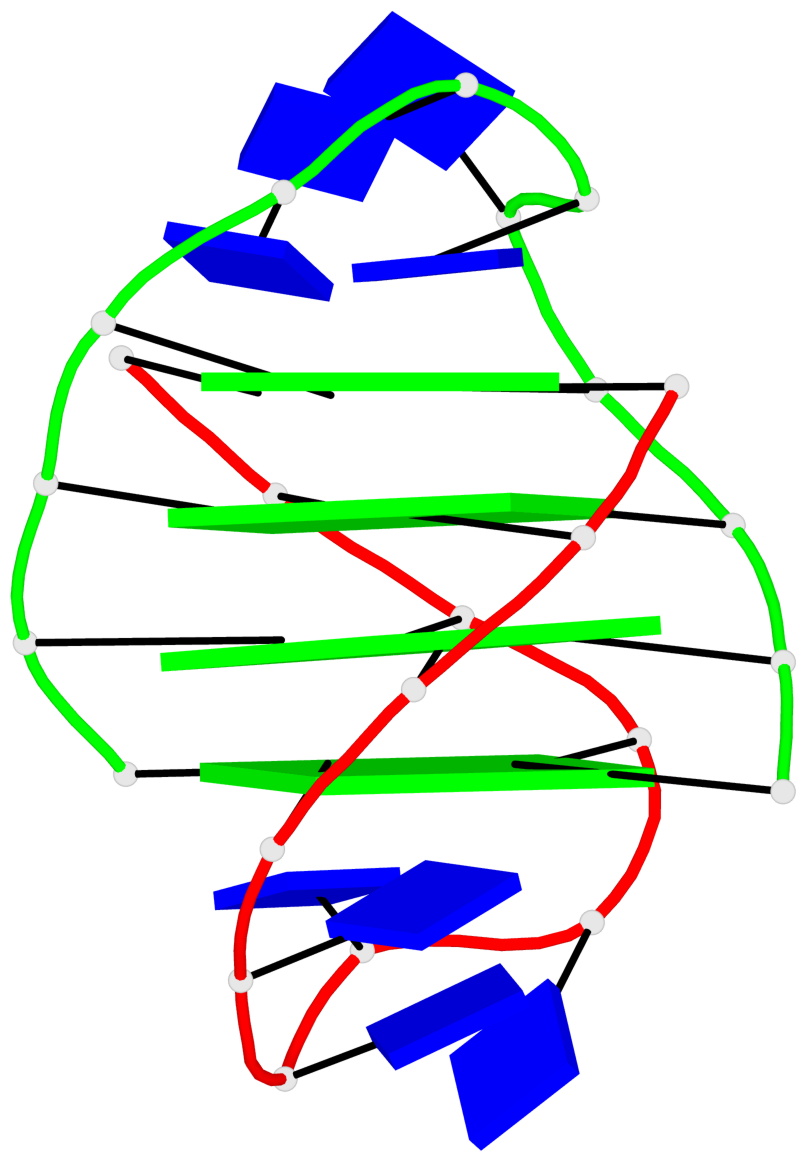
Detailed DSSR results for the G-quadruplex: PDB entry 2gwe
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 2gwe
- Class
- DNA
- Method
- X-ray (2.2 Å)
- Summary
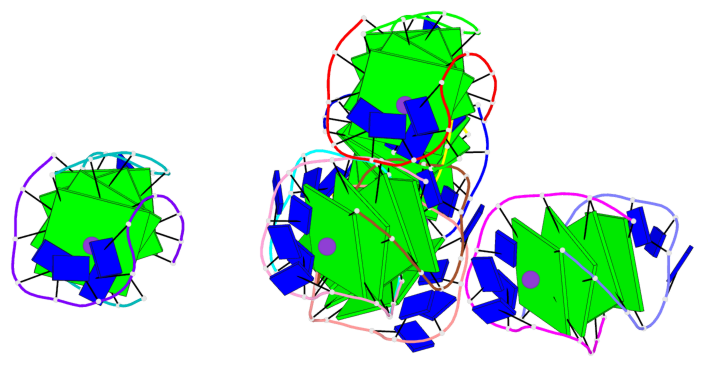
- Crystal structure of d(g4t4g4) with six quadruplexes in the asymmetric unit.
- Reference
- Lee MPH, Haider S, Parkinson GN, Neidle S: "Crystal structure of D(G4T4G4) with four and six quadruplexes in the asymmetric unit."
- Abstract
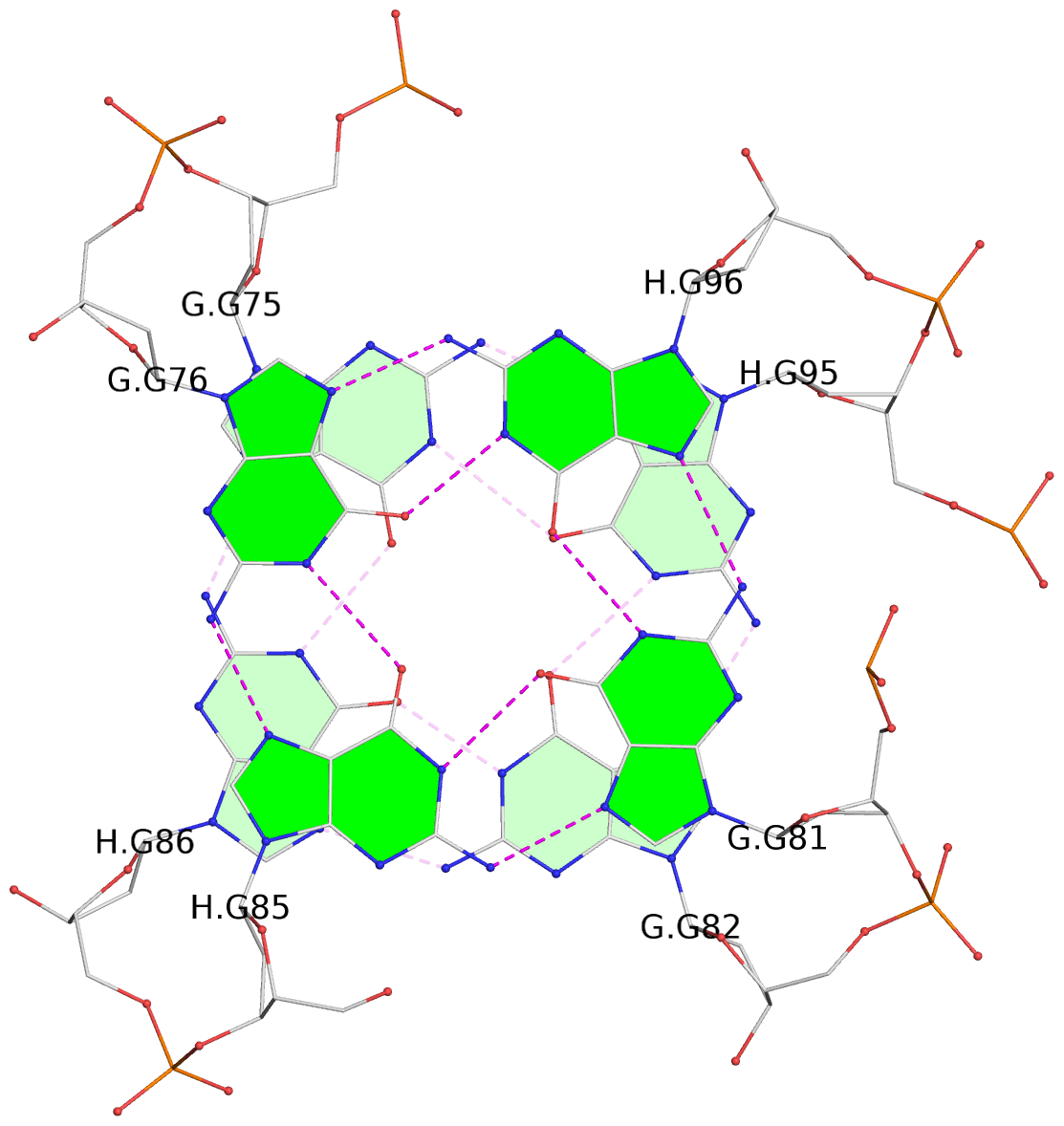
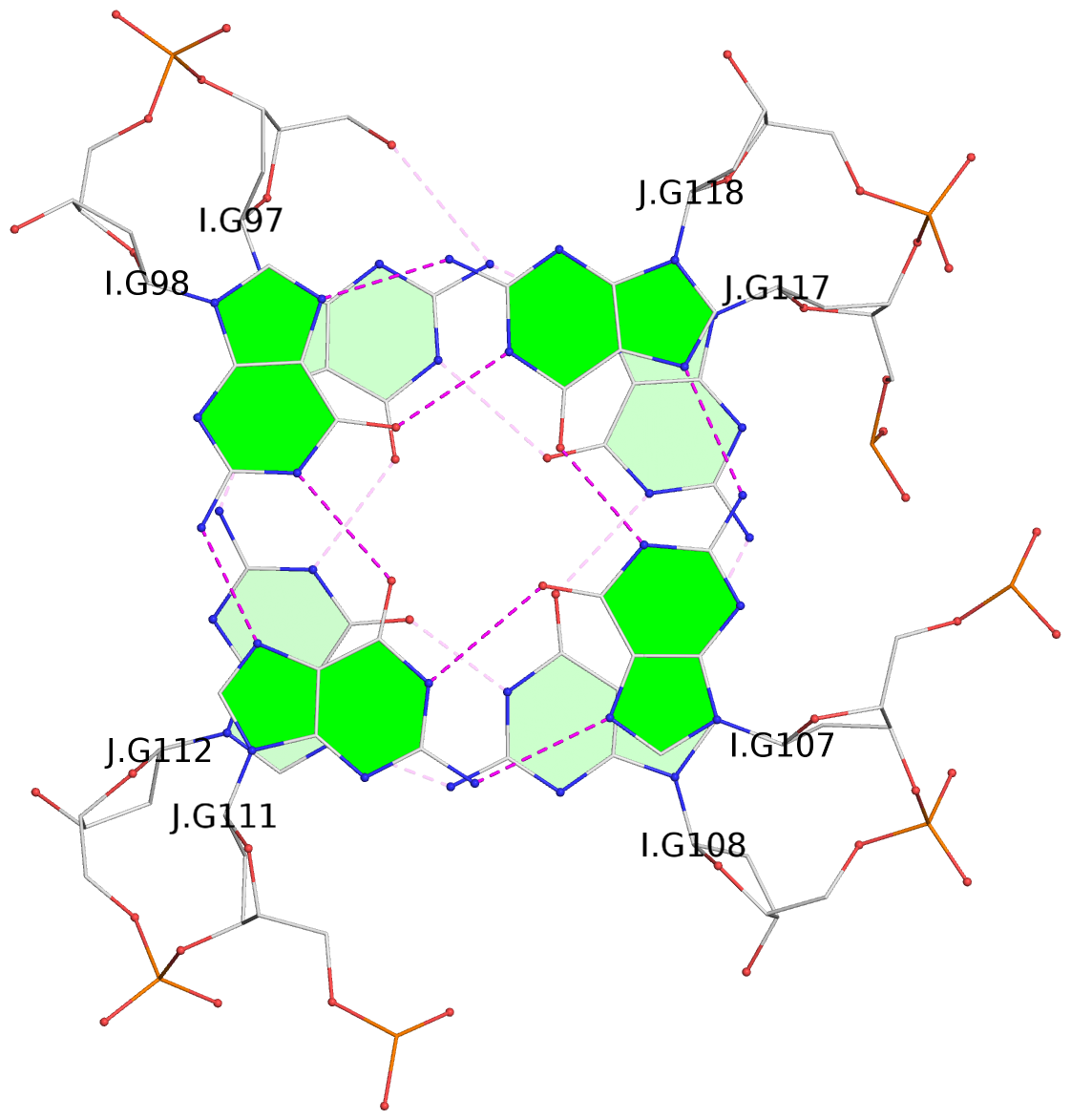
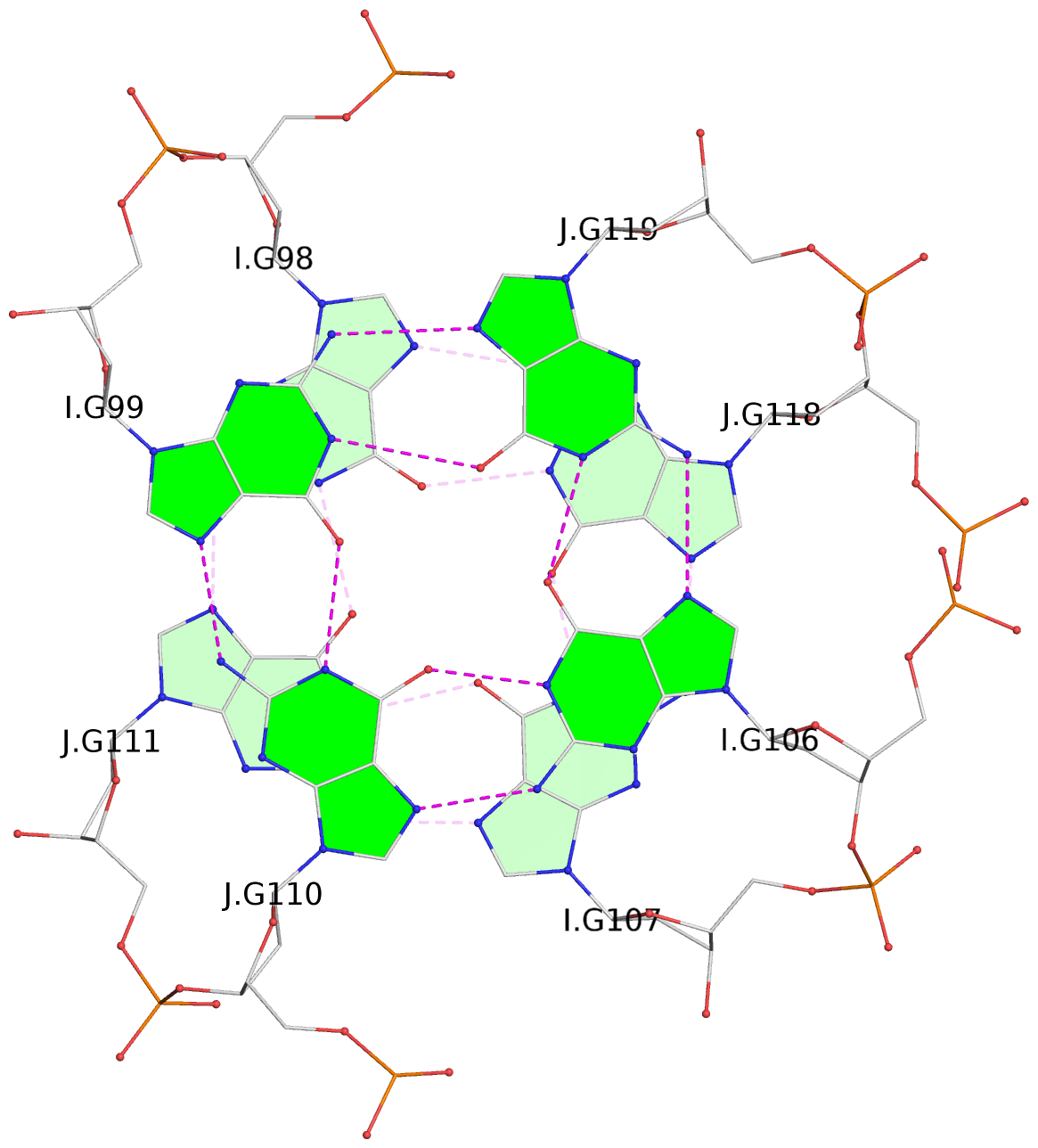
- G4 notes
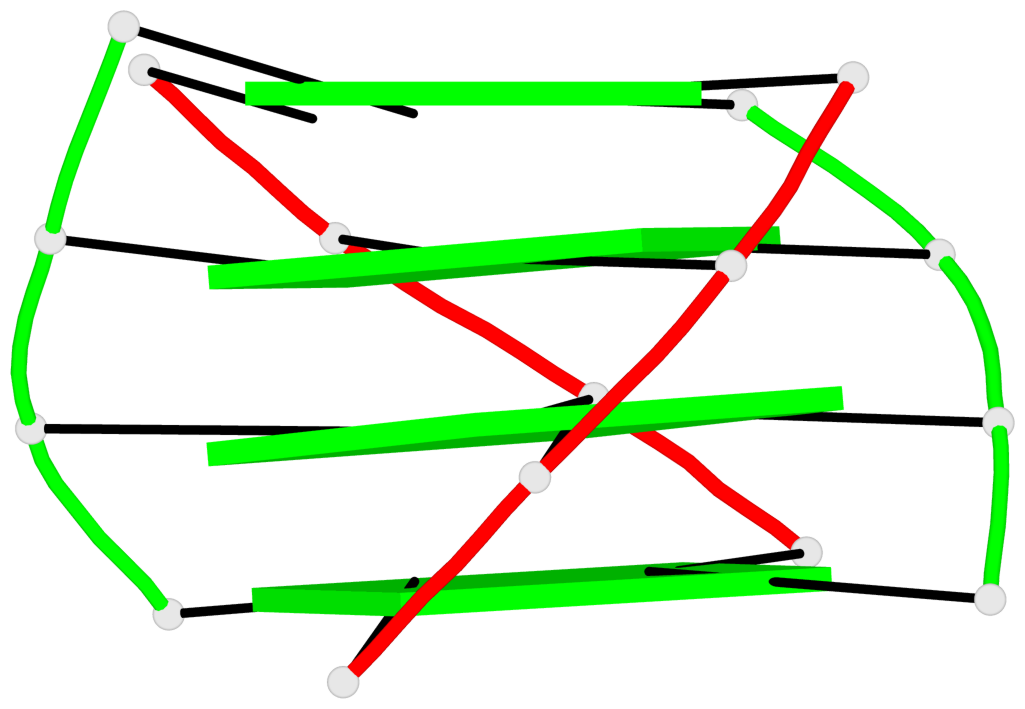
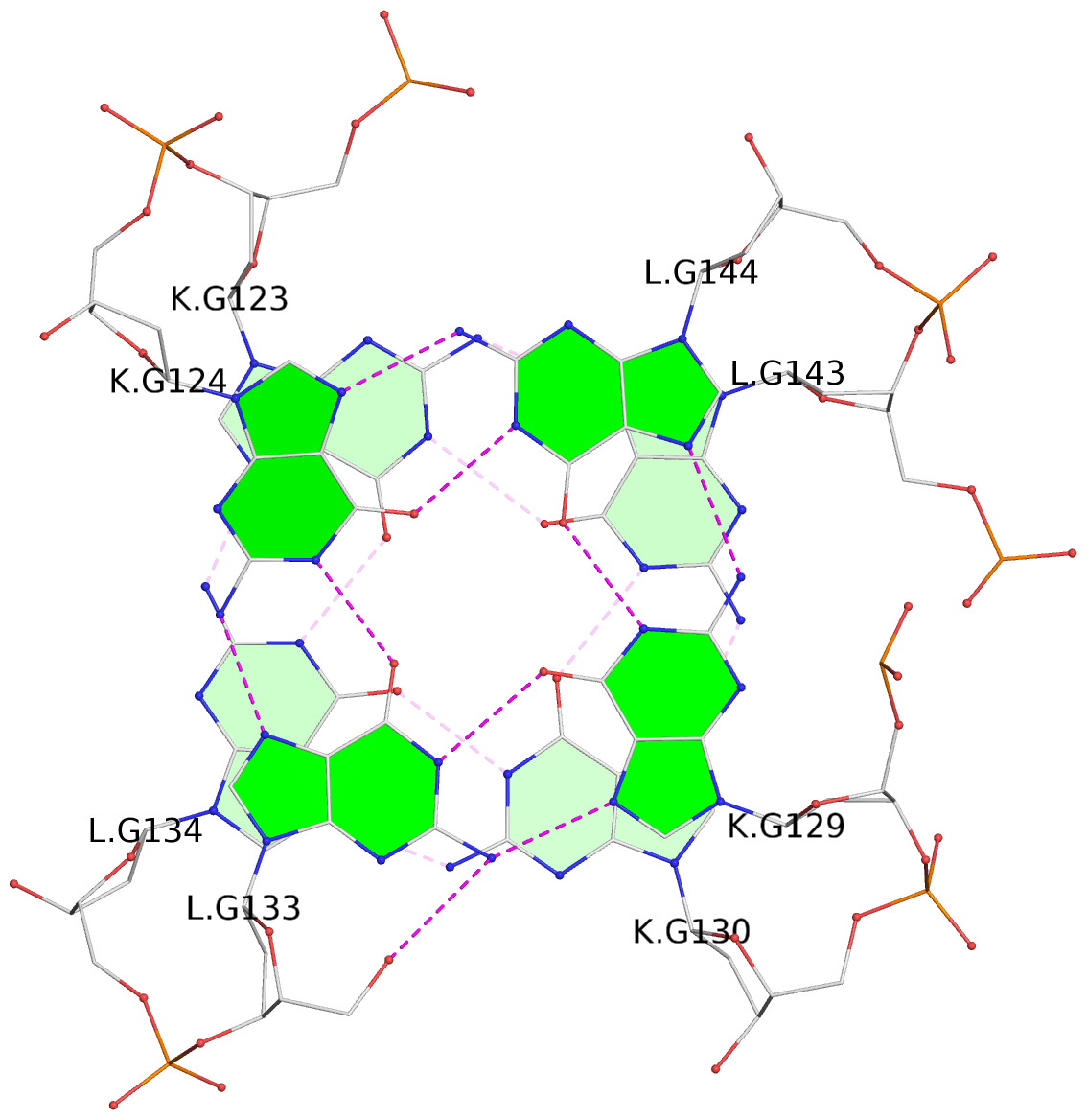
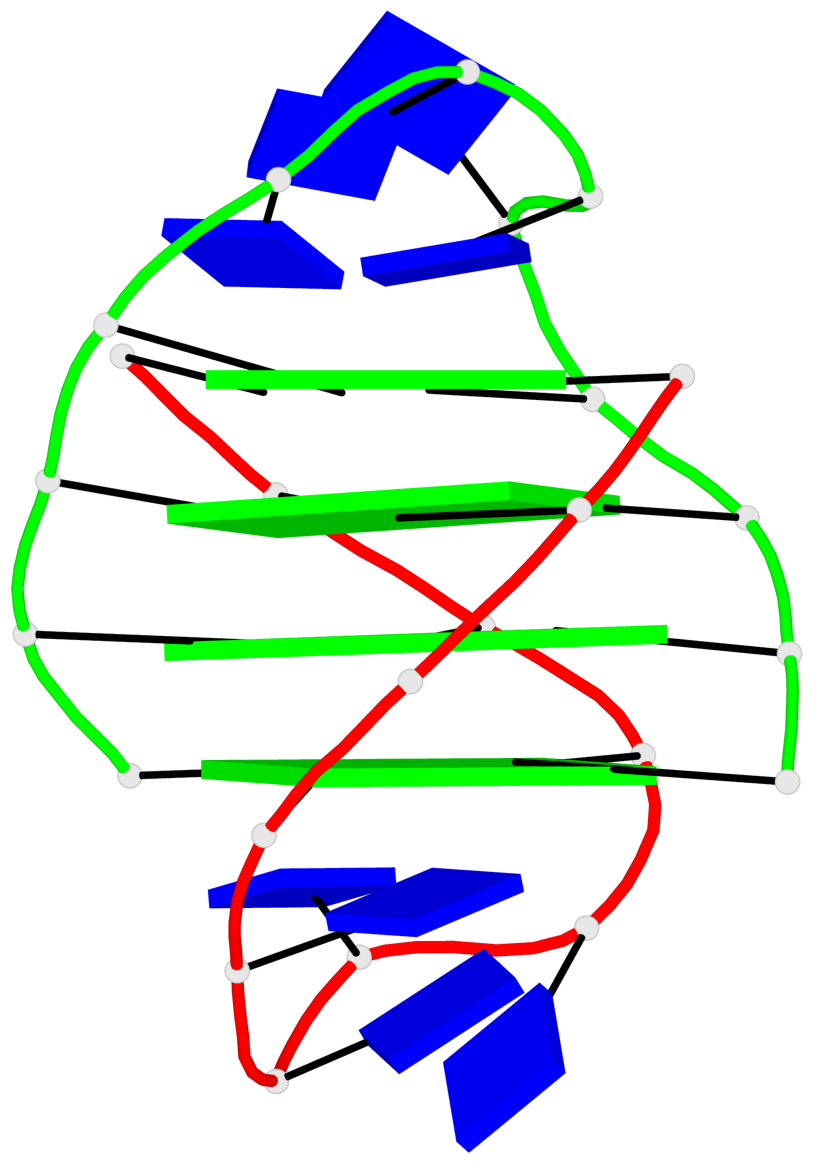
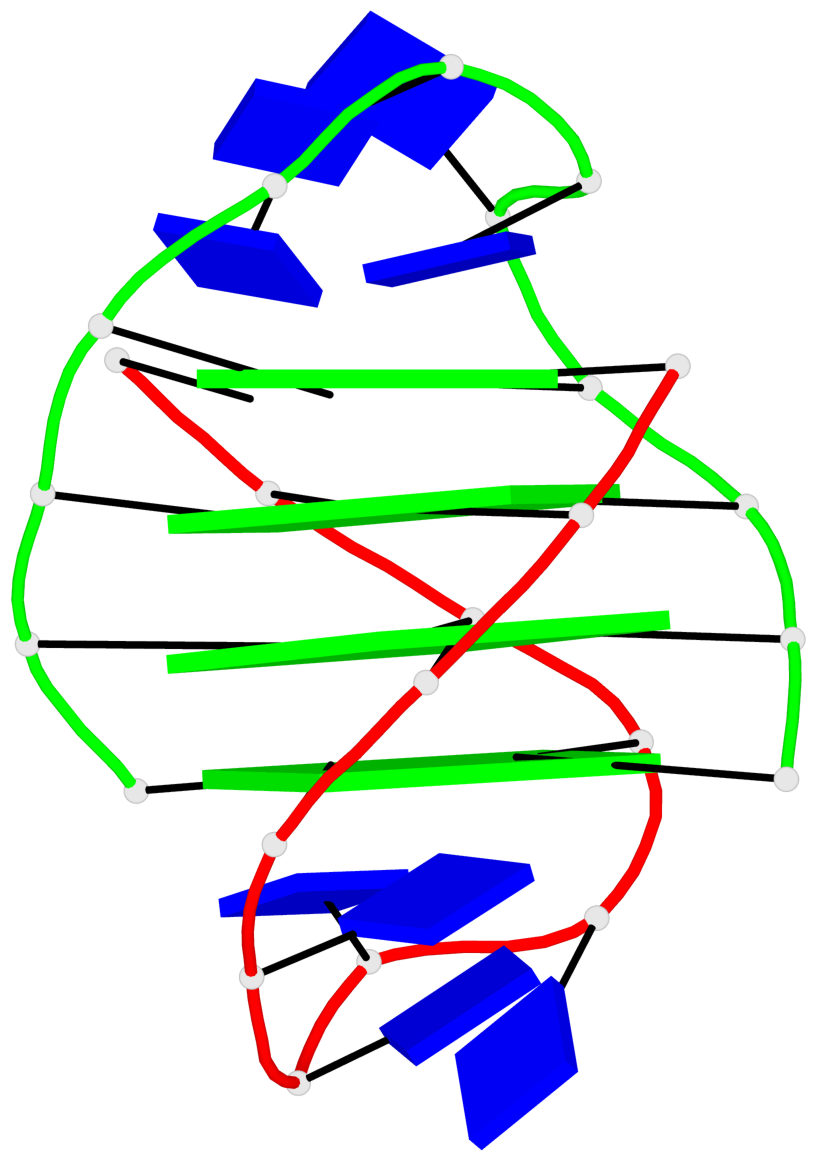
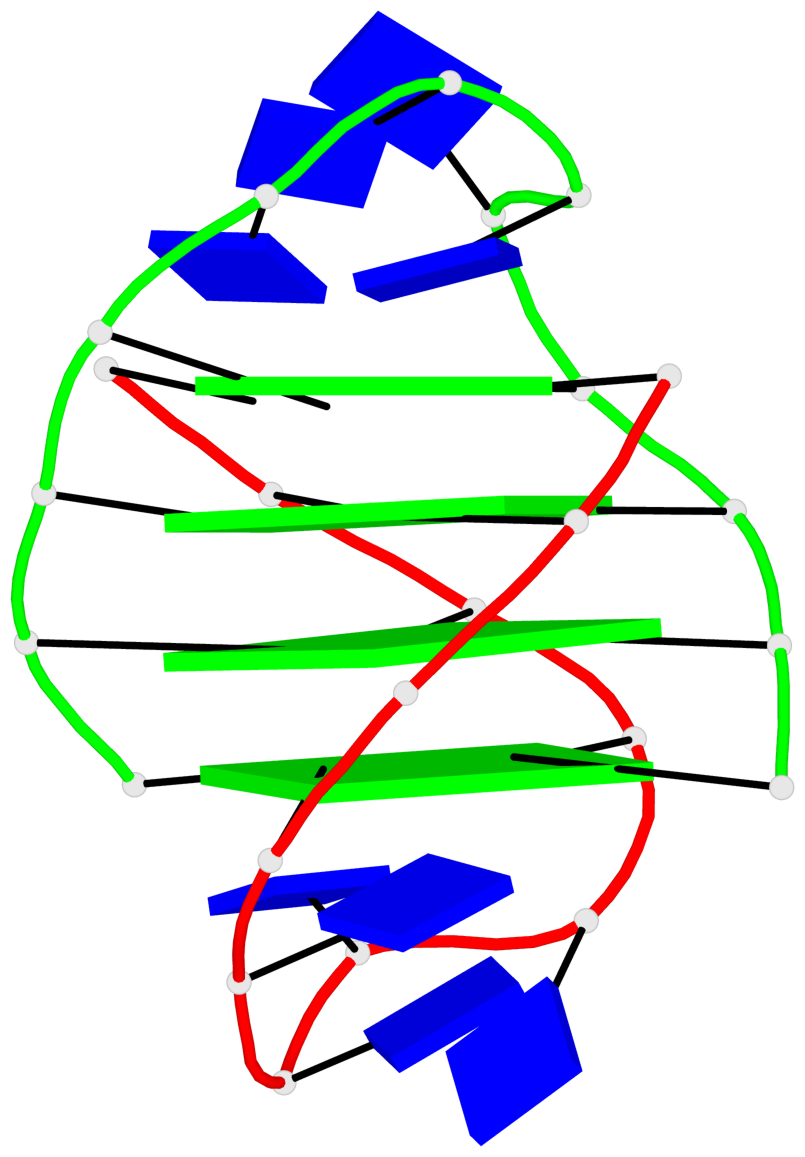
- 24 G-tetrads, 6 G4 helices, 6 G4 stems, (2+2), UDDU
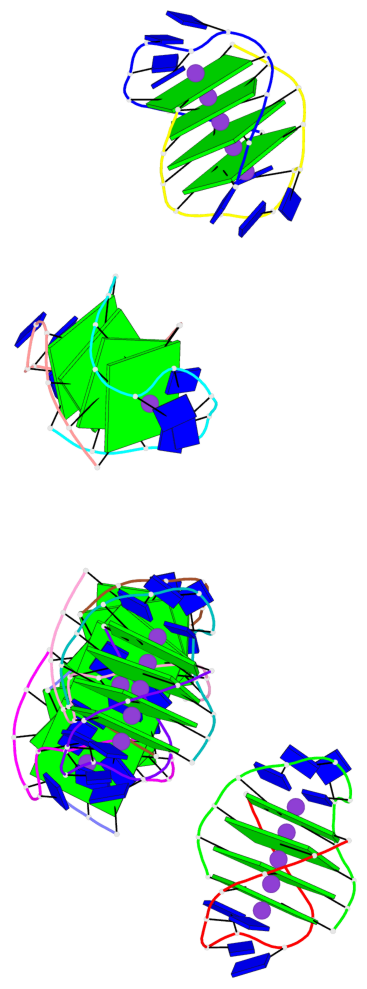
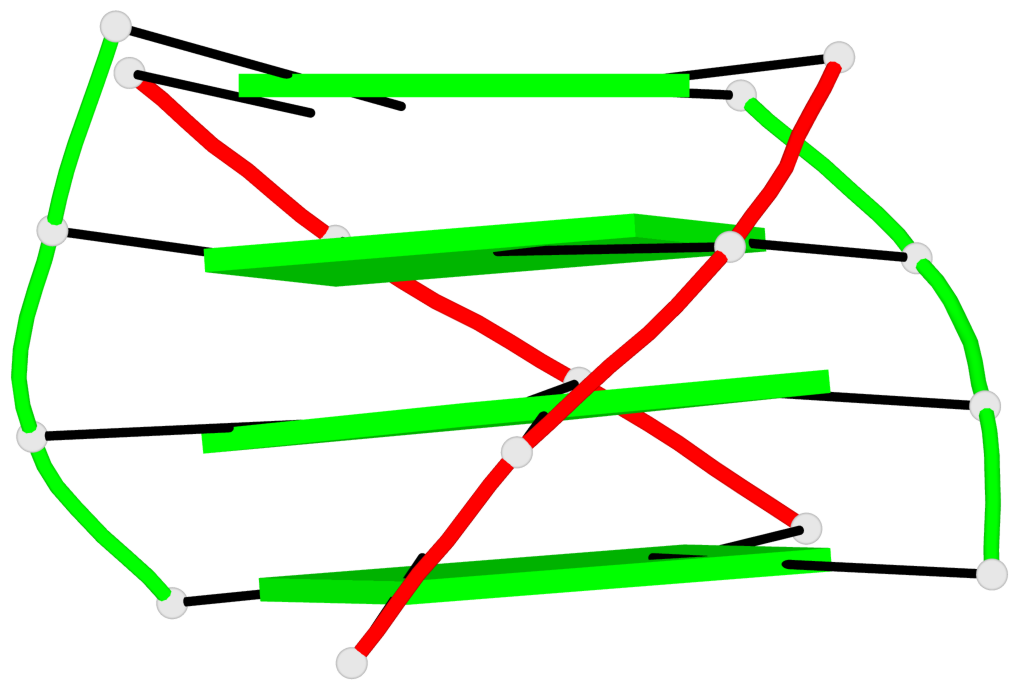
Base-block schematics in six views
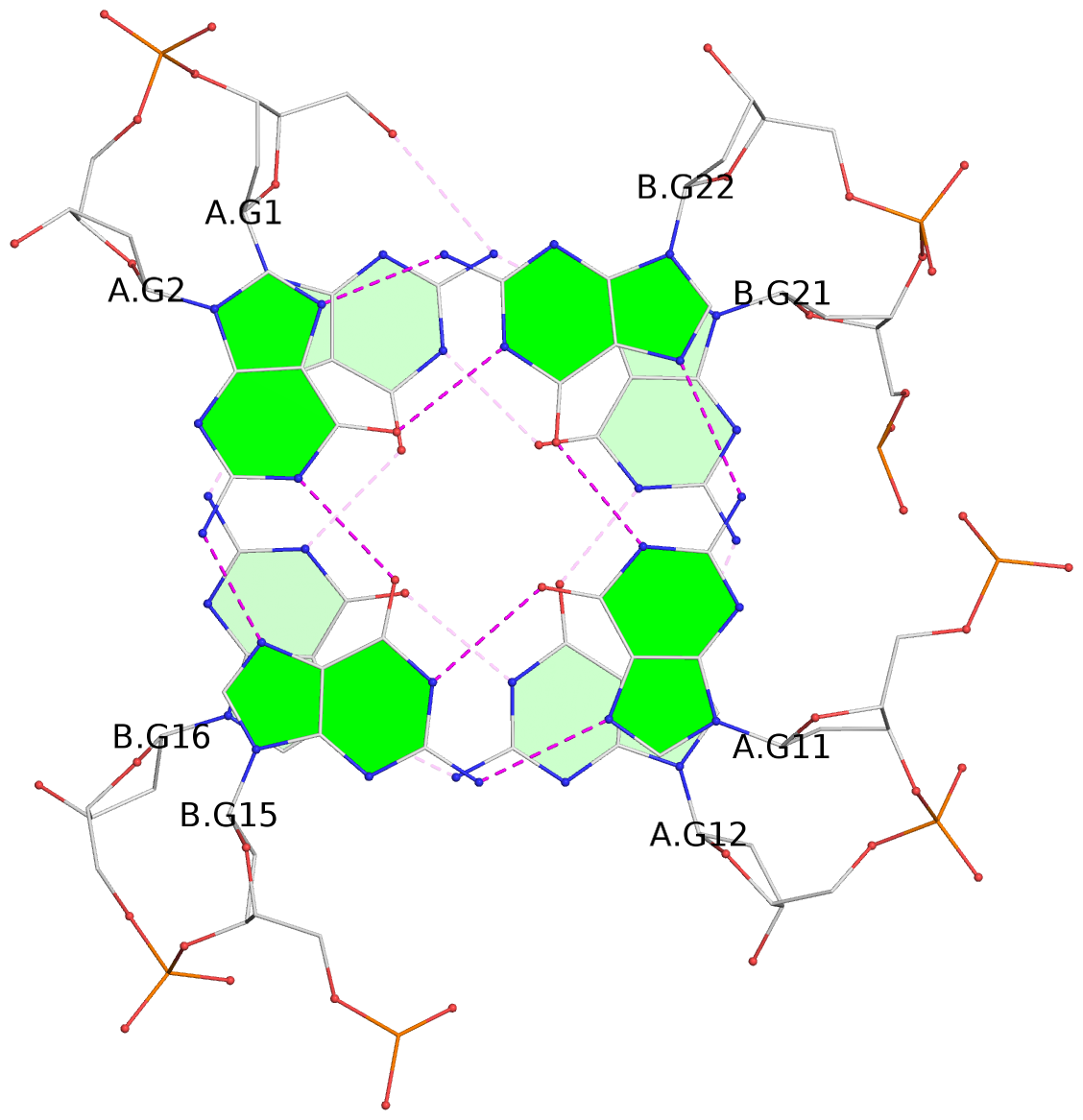
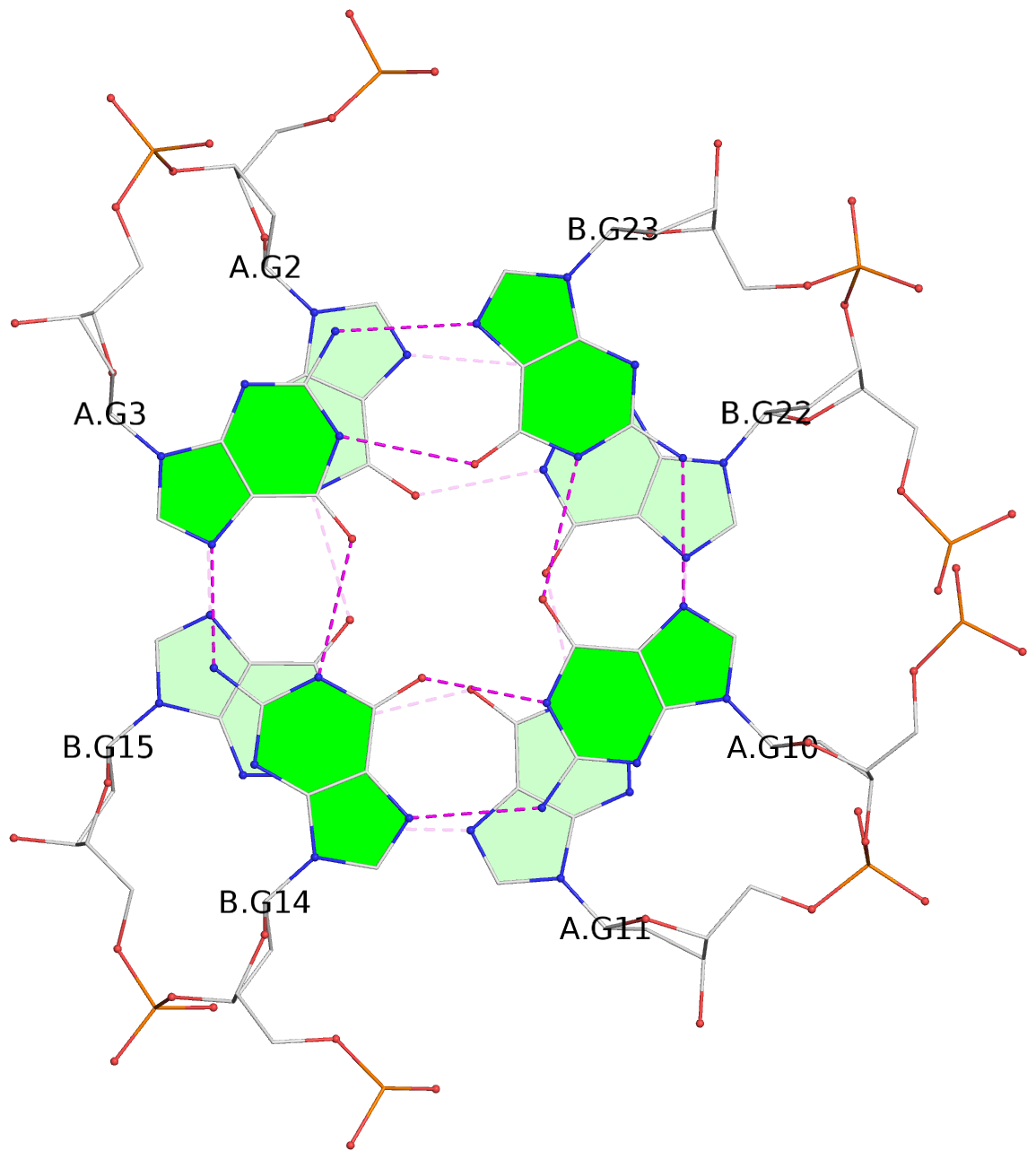
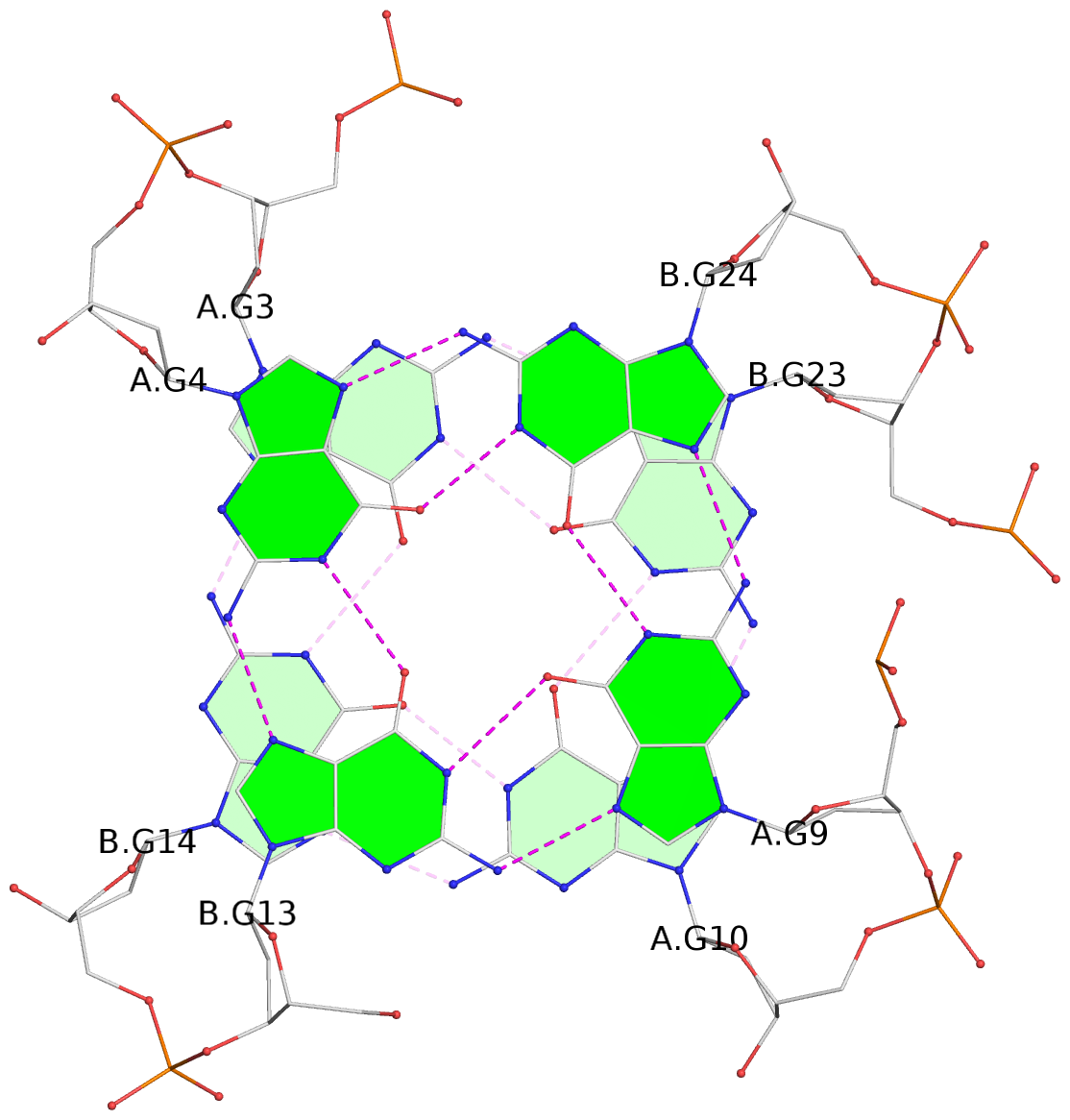
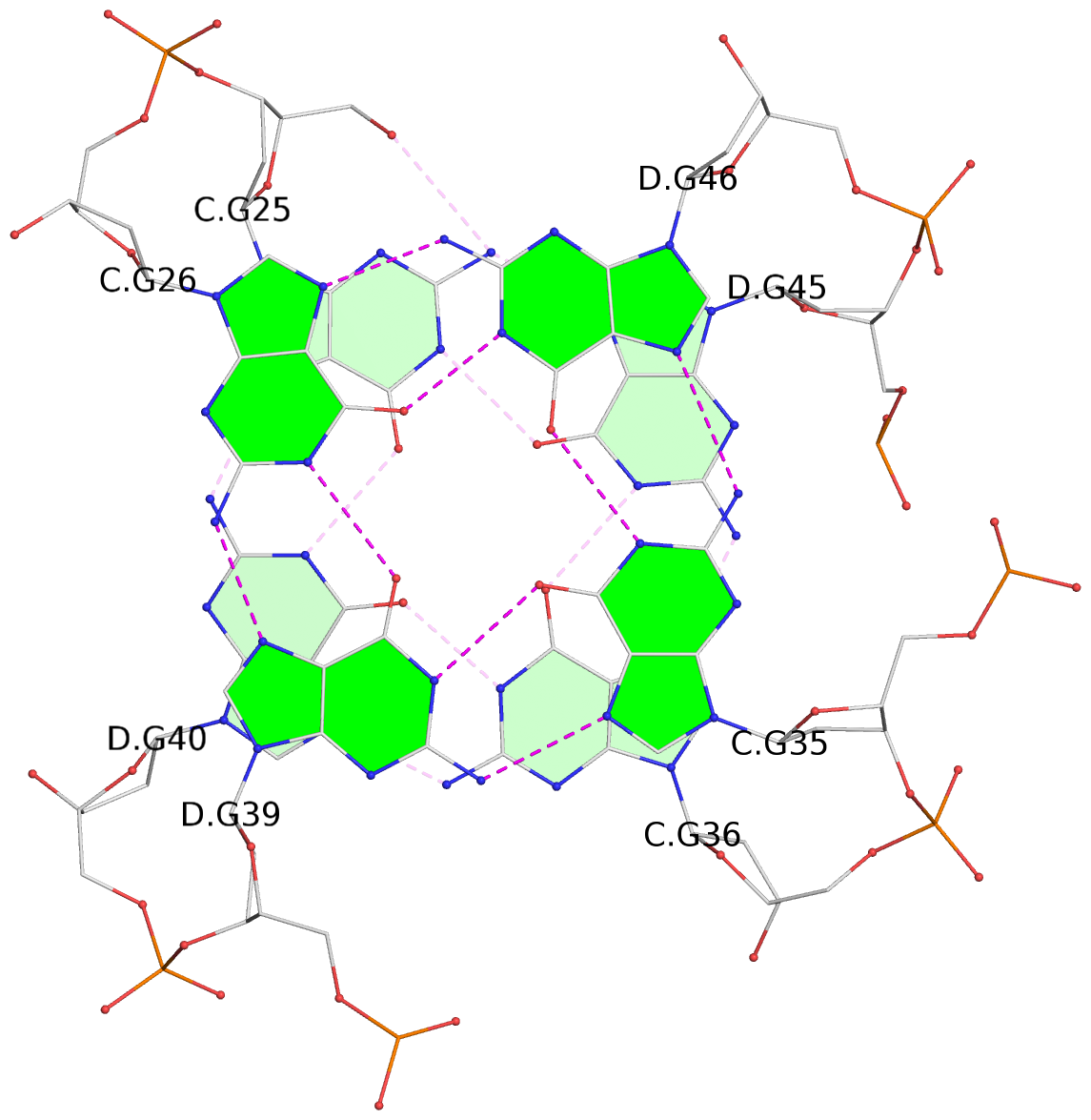
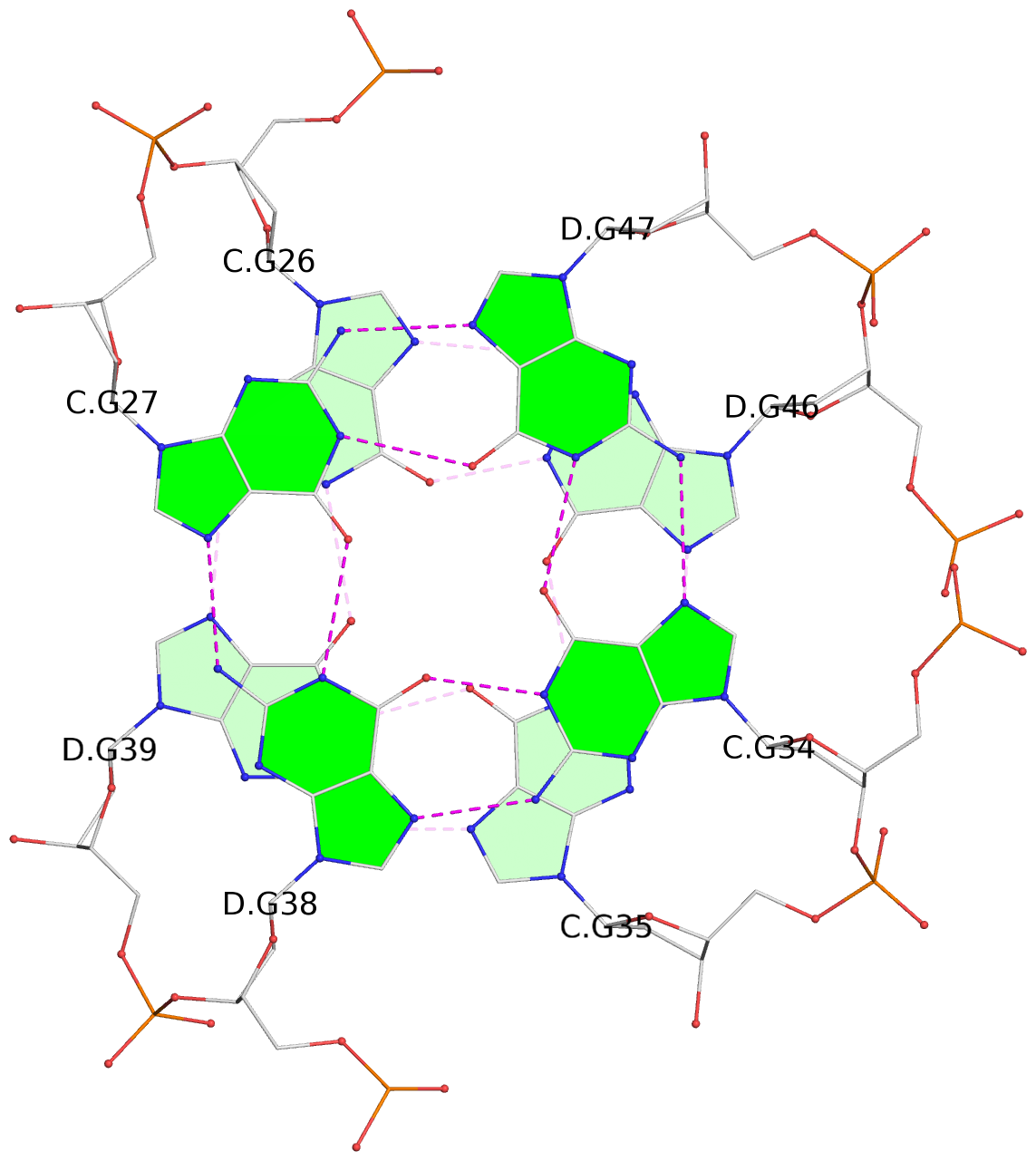
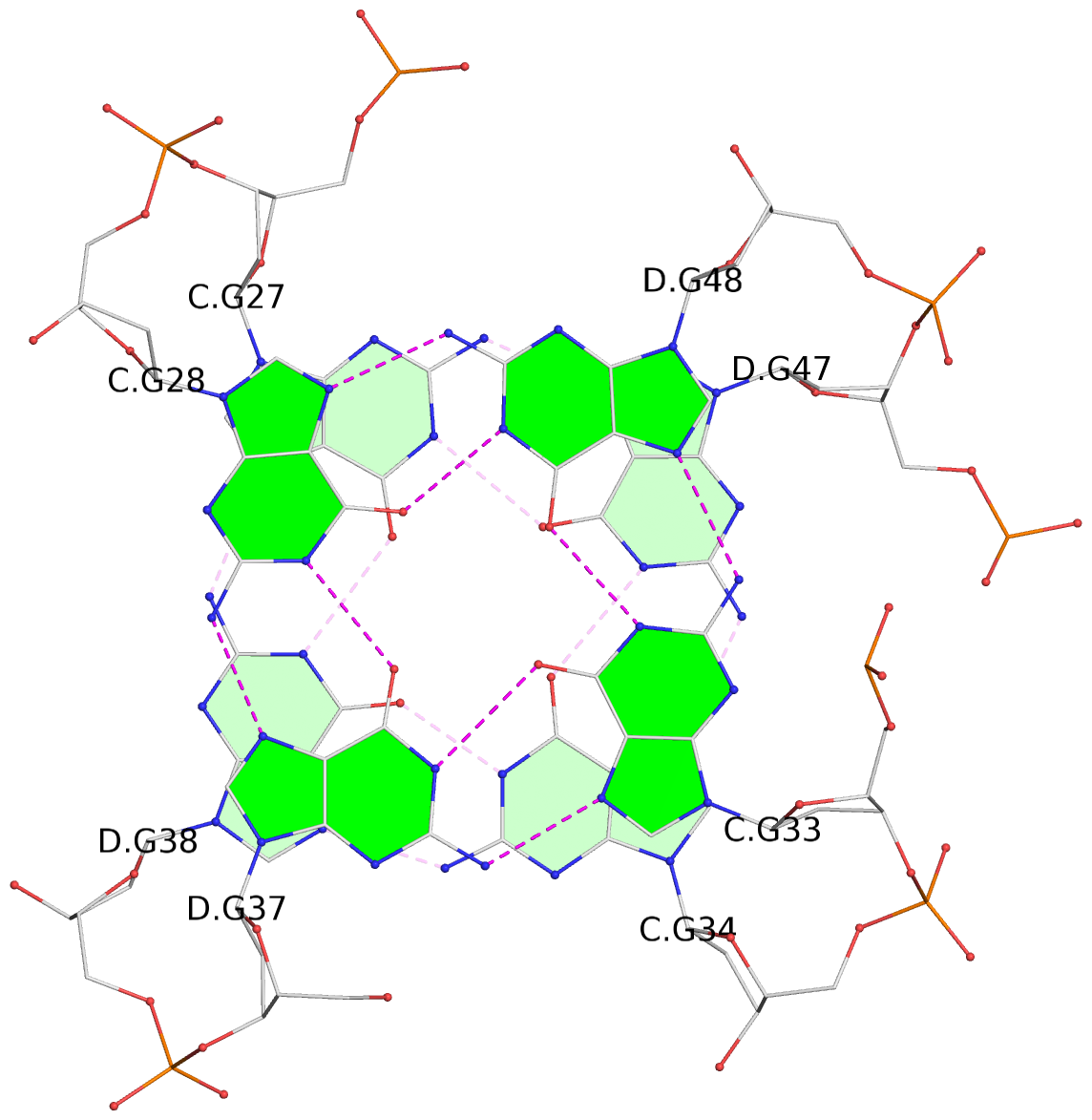
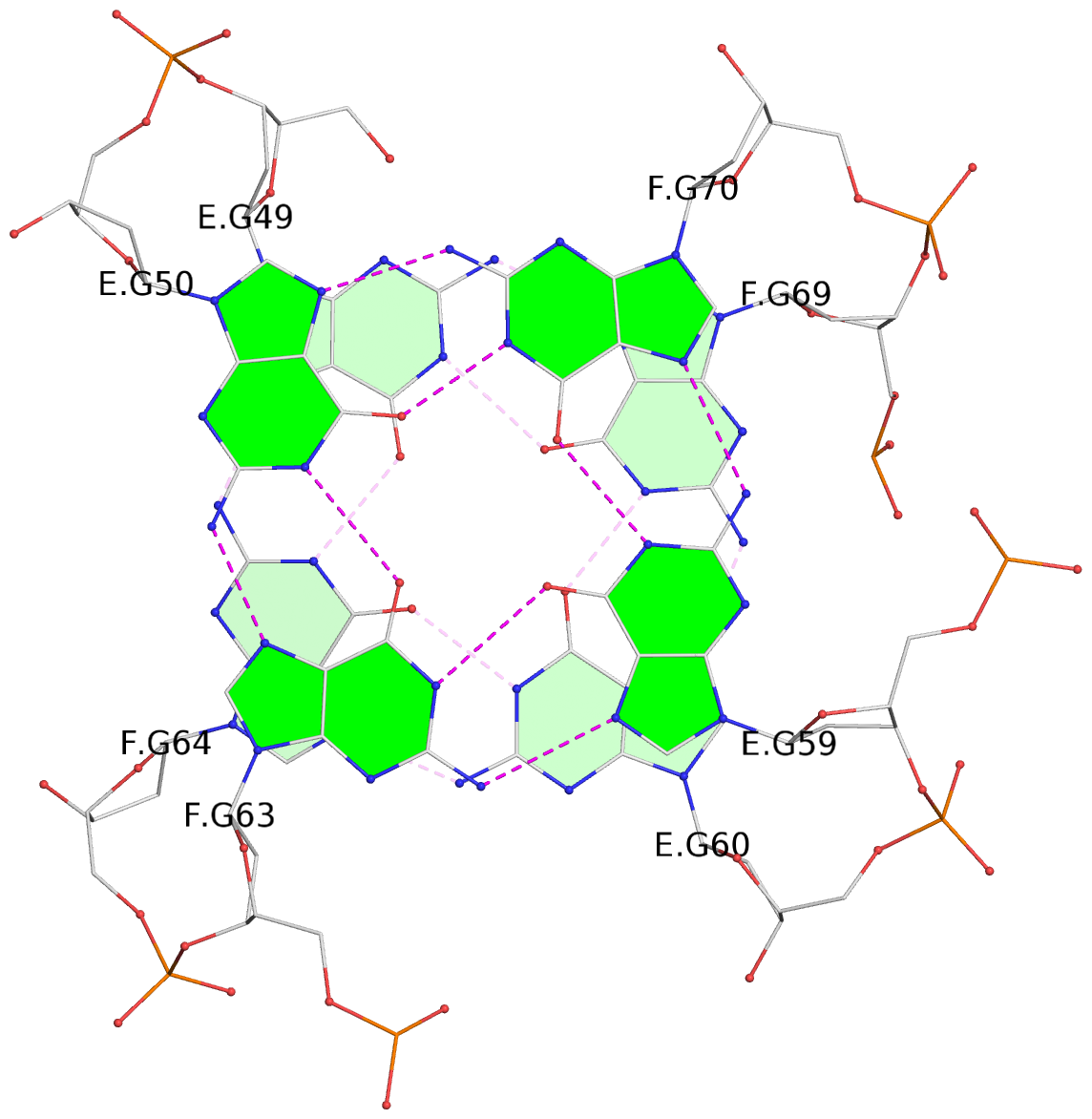
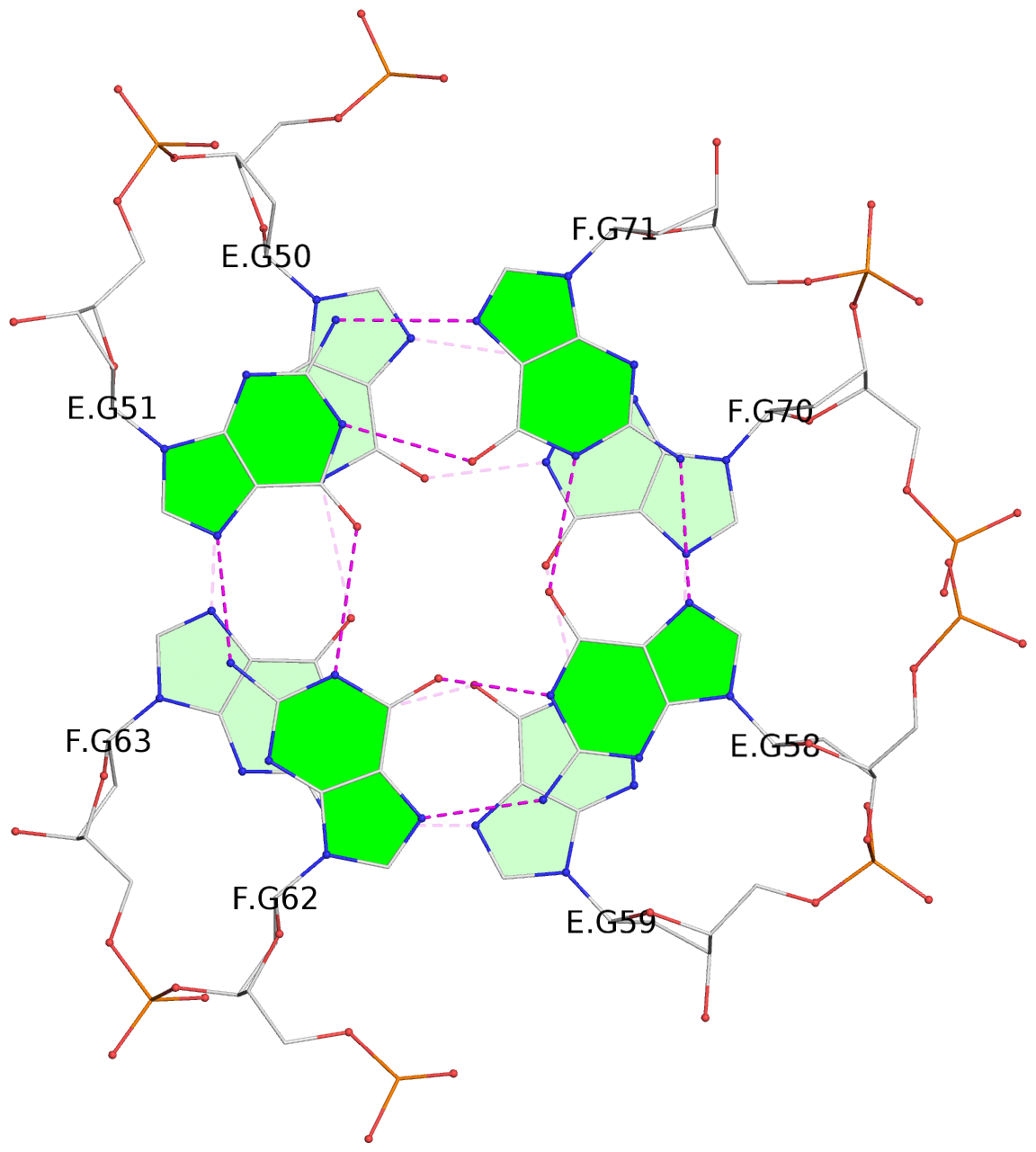
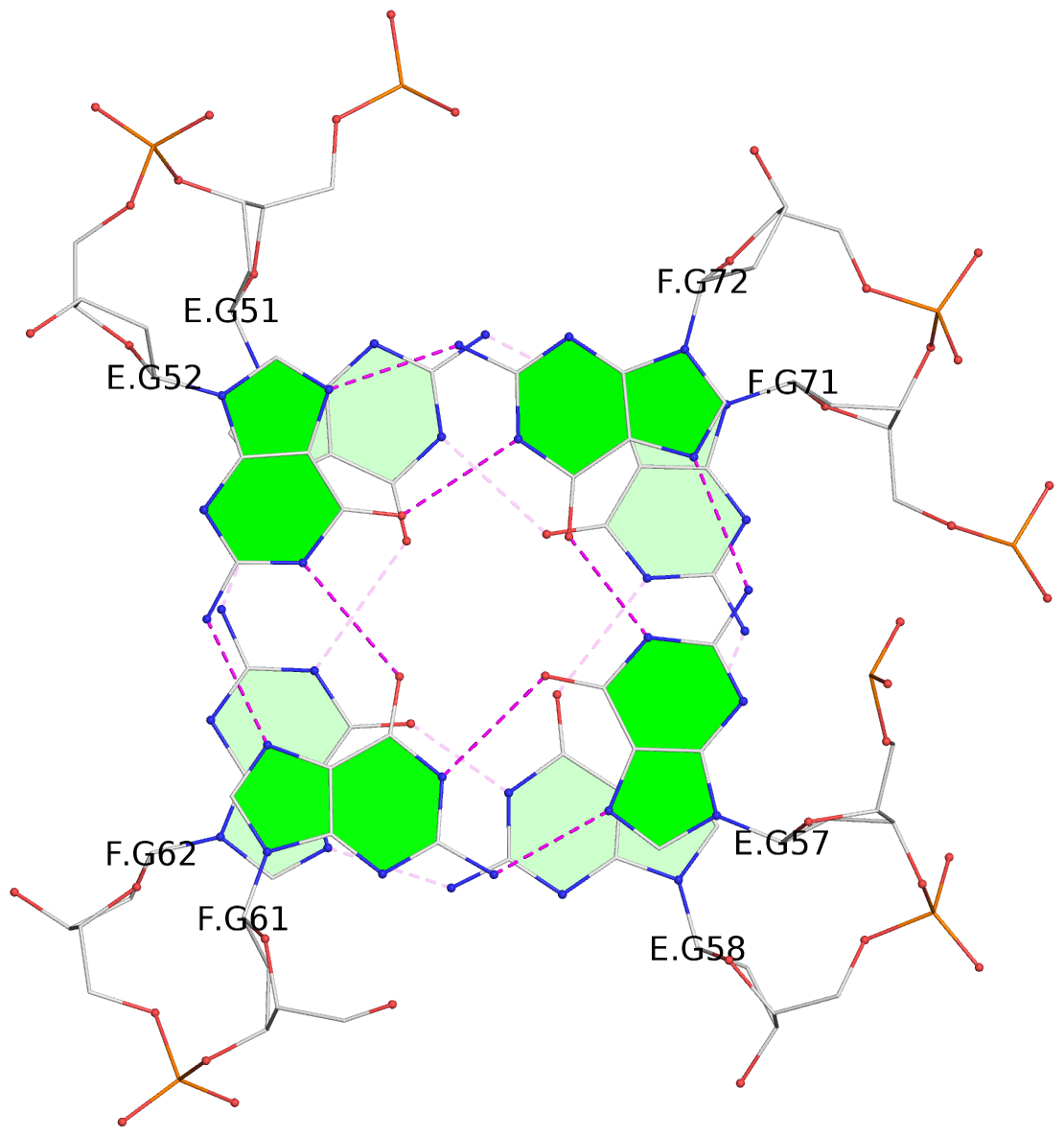
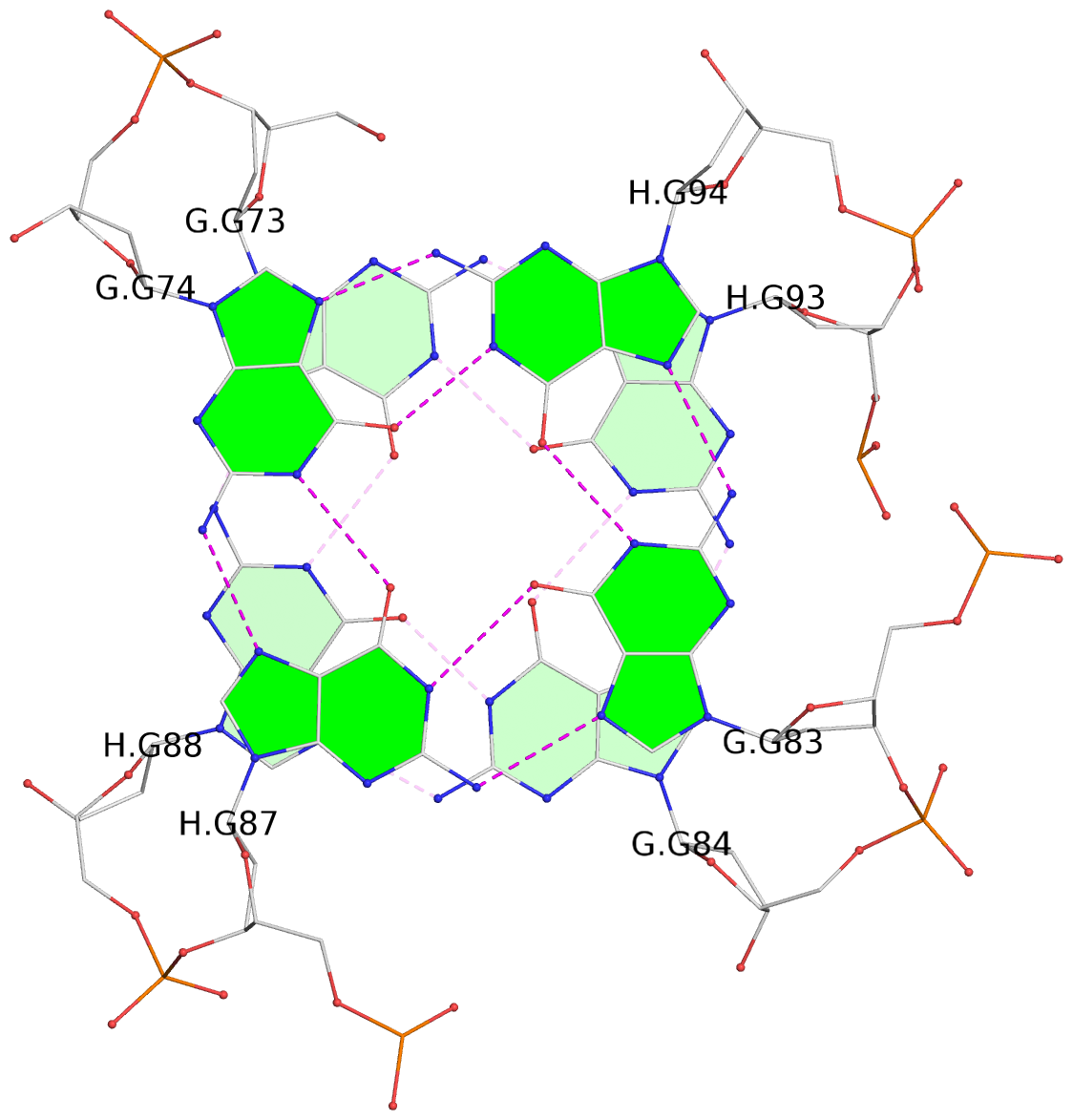
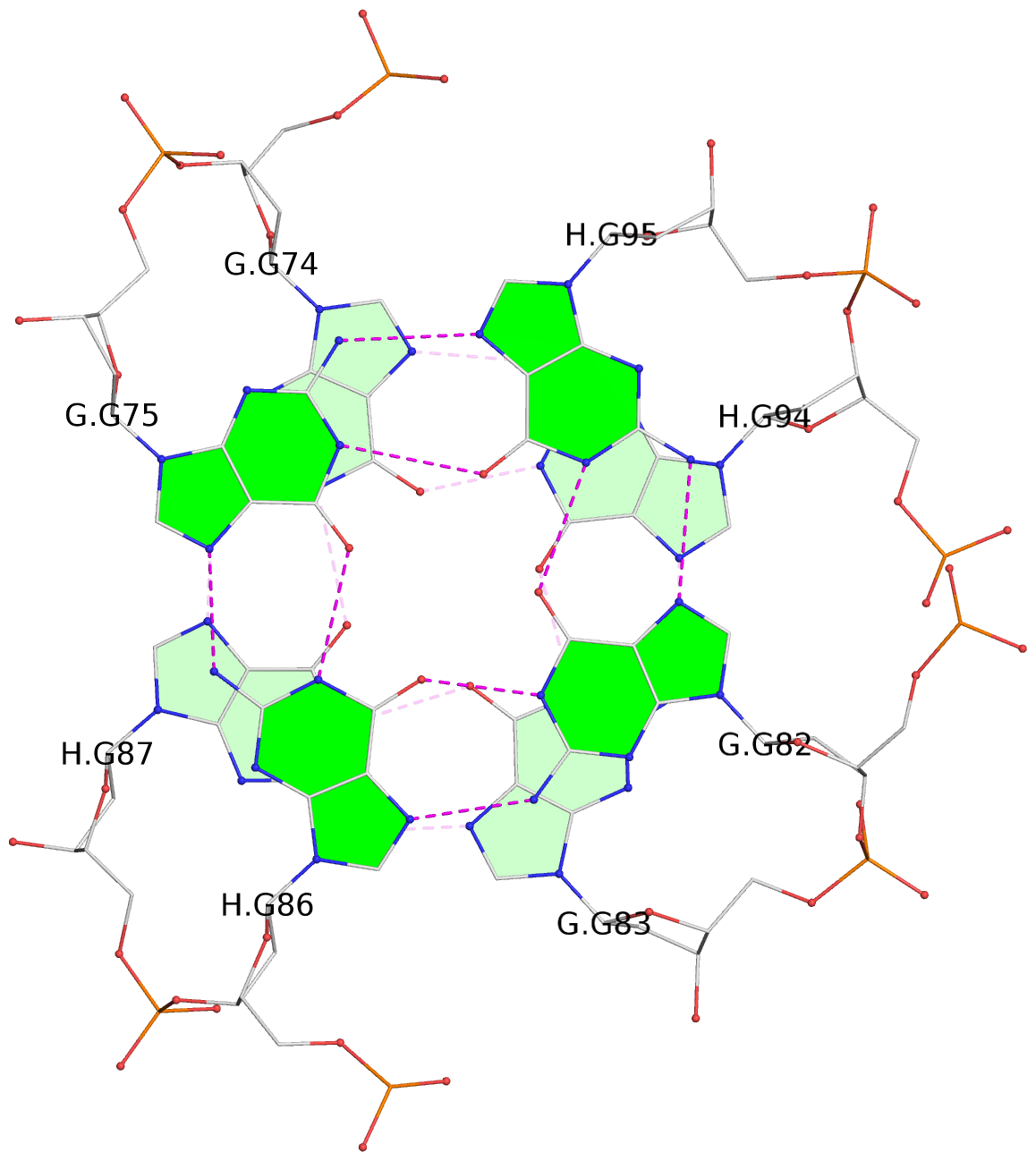
List of 24 G-tetrads
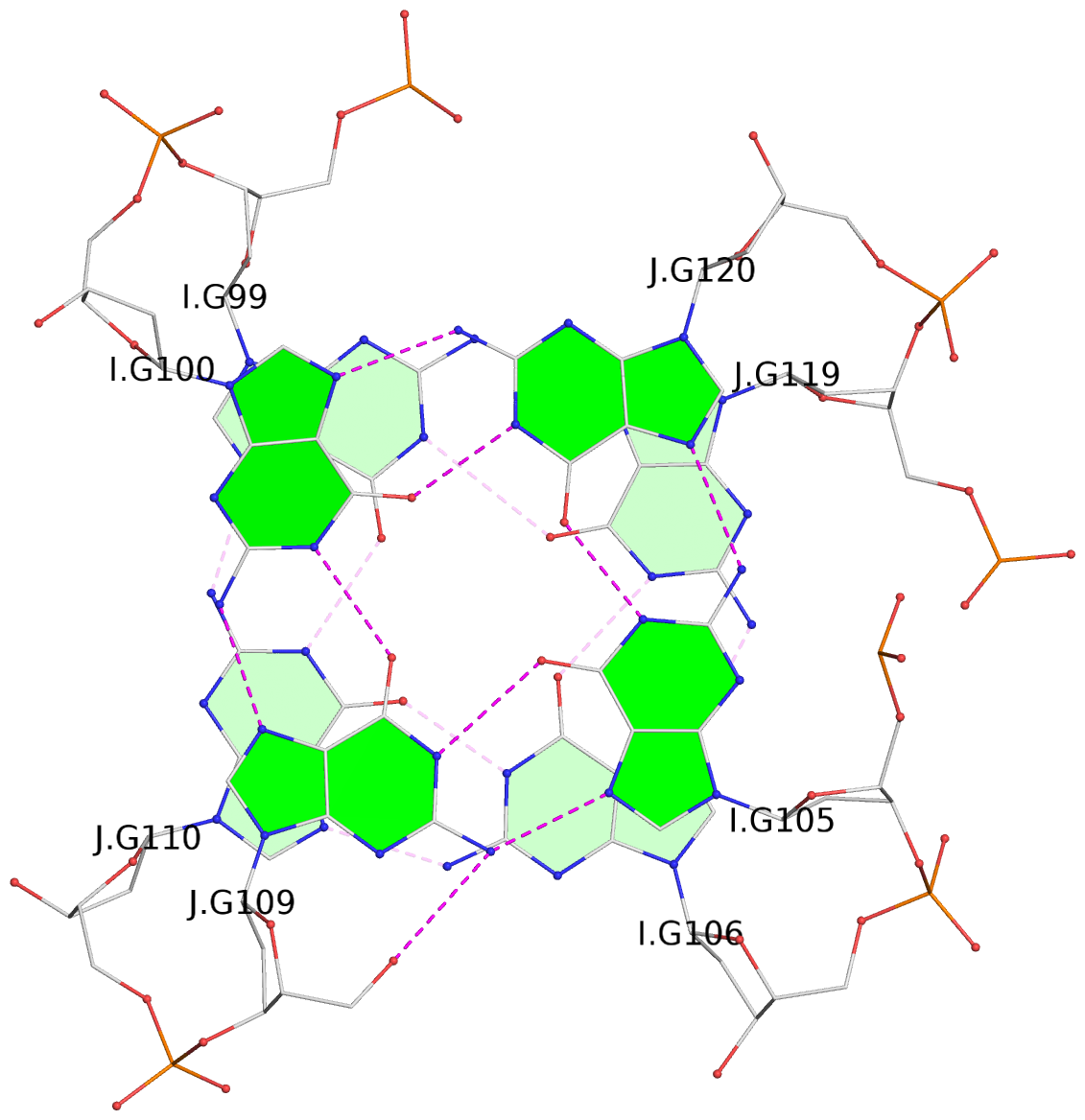
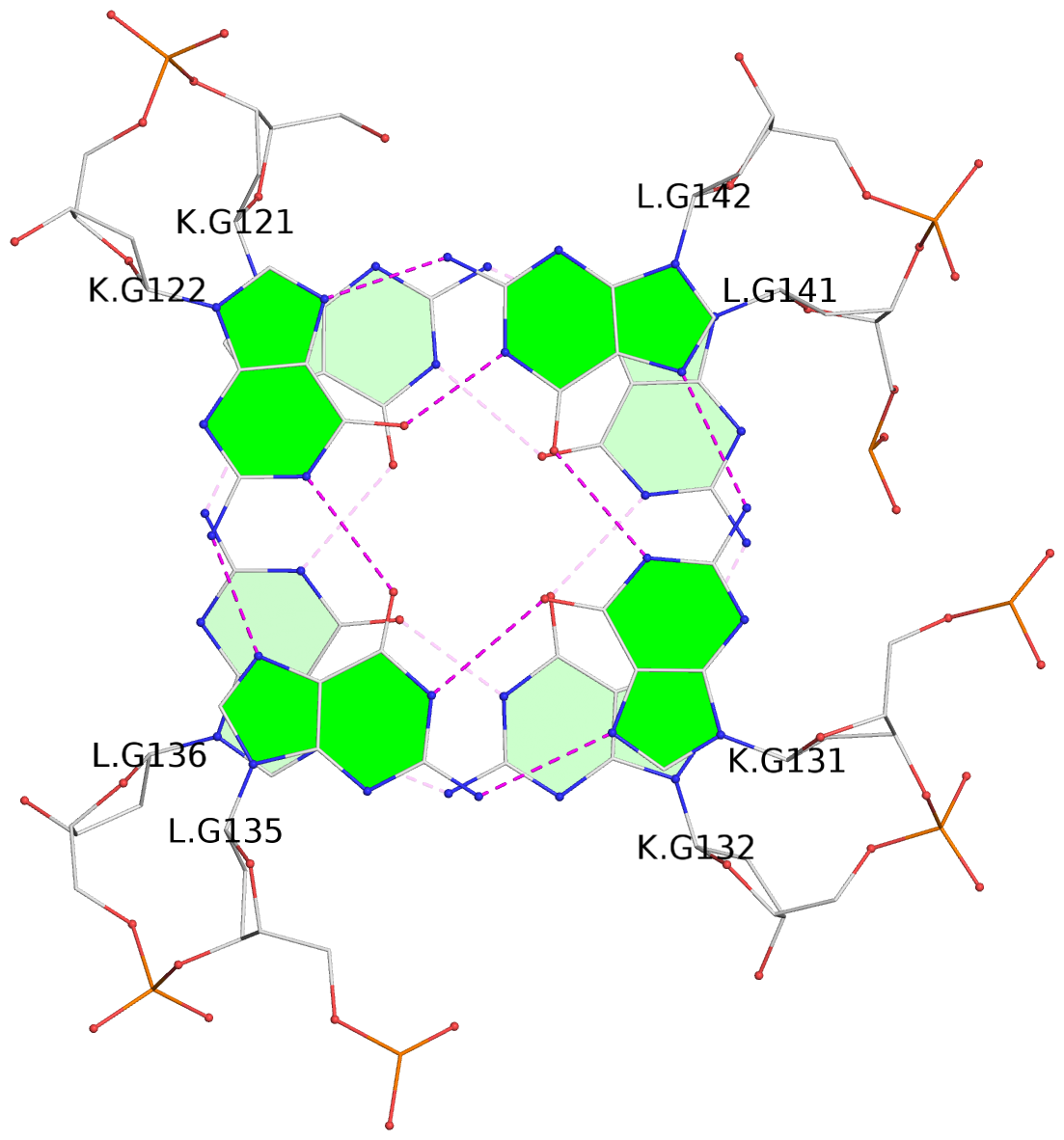
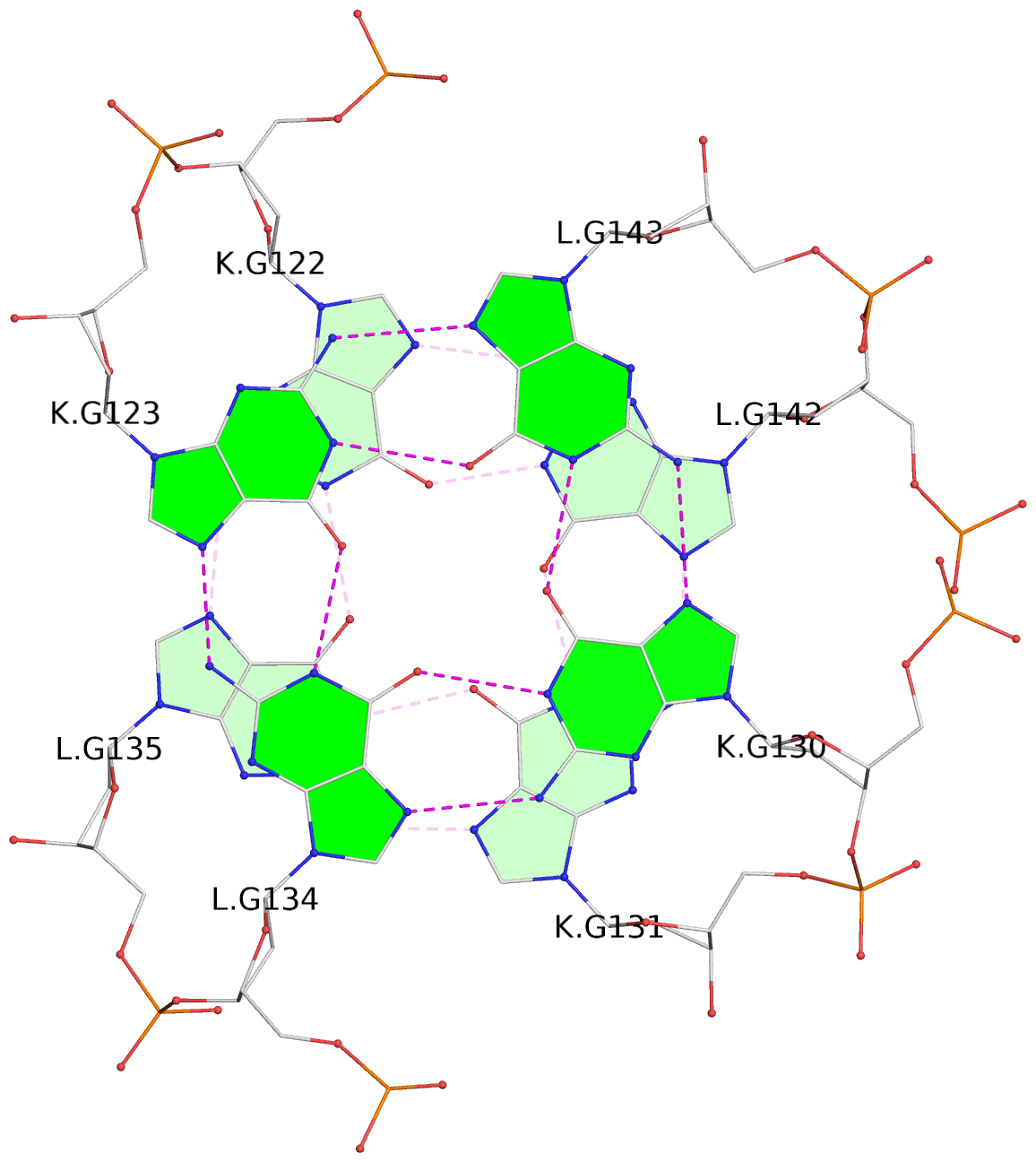
1 glyco-bond=s--s sugar=---- groove=w-n- planarity=0.267 type=bowl nts=4 GGGG A.DG1,B.DG16,A.DG12,B.DG21 2 glyco-bond=-ss- sugar=---- groove=w-n- planarity=0.164 type=other nts=4 GGGG A.DG2,B.DG15,A.DG11,B.DG22 3 glyco-bond=s--s sugar=---- groove=w-n- planarity=0.126 type=planar nts=4 GGGG A.DG3,B.DG14,A.DG10,B.DG23 4 glyco-bond=-ss- sugar=---- groove=w-n- planarity=0.250 type=other nts=4 GGGG A.DG4,B.DG13,A.DG9,B.DG24 5 glyco-bond=s--s sugar=---- groove=w-n- planarity=0.264 type=bowl nts=4 GGGG C.DG25,D.DG40,C.DG36,D.DG45 6 glyco-bond=-ss- sugar=---- groove=w-n- planarity=0.109 type=planar nts=4 GGGG C.DG26,D.DG39,C.DG35,D.DG46 7 glyco-bond=s--s sugar=---- groove=w-n- planarity=0.143 type=planar nts=4 GGGG C.DG27,D.DG38,C.DG34,D.DG47 8 glyco-bond=-ss- sugar=---- groove=w-n- planarity=0.245 type=other nts=4 GGGG C.DG28,D.DG37,C.DG33,D.DG48 9 glyco-bond=s--s sugar=---- groove=w-n- planarity=0.291 type=bowl nts=4 GGGG E.DG49,F.DG64,E.DG60,F.DG69 10 glyco-bond=-ss- sugar=---- groove=w-n- planarity=0.105 type=planar nts=4 GGGG E.DG50,F.DG63,E.DG59,F.DG70 11 glyco-bond=s--s sugar=---- groove=w-n- planarity=0.100 type=planar nts=4 GGGG E.DG51,F.DG62,E.DG58,F.DG71 12 glyco-bond=-ss- sugar=---- groove=w-n- planarity=0.274 type=bowl nts=4 GGGG E.DG52,F.DG61,E.DG57,F.DG72 13 glyco-bond=s--s sugar=---- groove=w-n- planarity=0.272 type=other nts=4 GGGG G.DG73,H.DG88,G.DG84,H.DG93 14 glyco-bond=-ss- sugar=---- groove=w-n- planarity=0.193 type=other nts=4 GGGG G.DG74,H.DG87,G.DG83,H.DG94 15 glyco-bond=s--s sugar=---- groove=w-n- planarity=0.169 type=other nts=4 GGGG G.DG75,H.DG86,G.DG82,H.DG95 16 glyco-bond=-ss- sugar=---- groove=w-n- planarity=0.256 type=other nts=4 GGGG G.DG76,H.DG85,G.DG81,H.DG96 17 glyco-bond=s--s sugar=---- groove=w-n- planarity=0.282 type=bowl nts=4 GGGG I.DG97,J.DG112,I.DG108,J.DG117 18 glyco-bond=-ss- sugar=---- groove=w-n- planarity=0.099 type=planar nts=4 GGGG I.DG98,J.DG111,I.DG107,J.DG118 19 glyco-bond=s--s sugar=---- groove=w-n- planarity=0.118 type=planar nts=4 GGGG I.DG99,J.DG110,I.DG106,J.DG119 20 glyco-bond=-ss- sugar=---- groove=w-n- planarity=0.281 type=bowl nts=4 GGGG I.DG100,J.DG109,I.DG105,J.DG120 21 glyco-bond=s--s sugar=---- groove=w-n- planarity=0.269 type=bowl nts=4 GGGG K.DG121,L.DG136,K.DG132,L.DG141 22 glyco-bond=-ss- sugar=---- groove=w-n- planarity=0.156 type=planar nts=4 GGGG K.DG122,L.DG135,K.DG131,L.DG142 23 glyco-bond=s--s sugar=---- groove=w-n- planarity=0.163 type=other nts=4 GGGG K.DG123,L.DG134,K.DG130,L.DG143 24 glyco-bond=-ss- sugar=---- groove=w-n- planarity=0.294 type=other nts=4 GGGG K.DG124,L.DG133,K.DG129,L.DG144
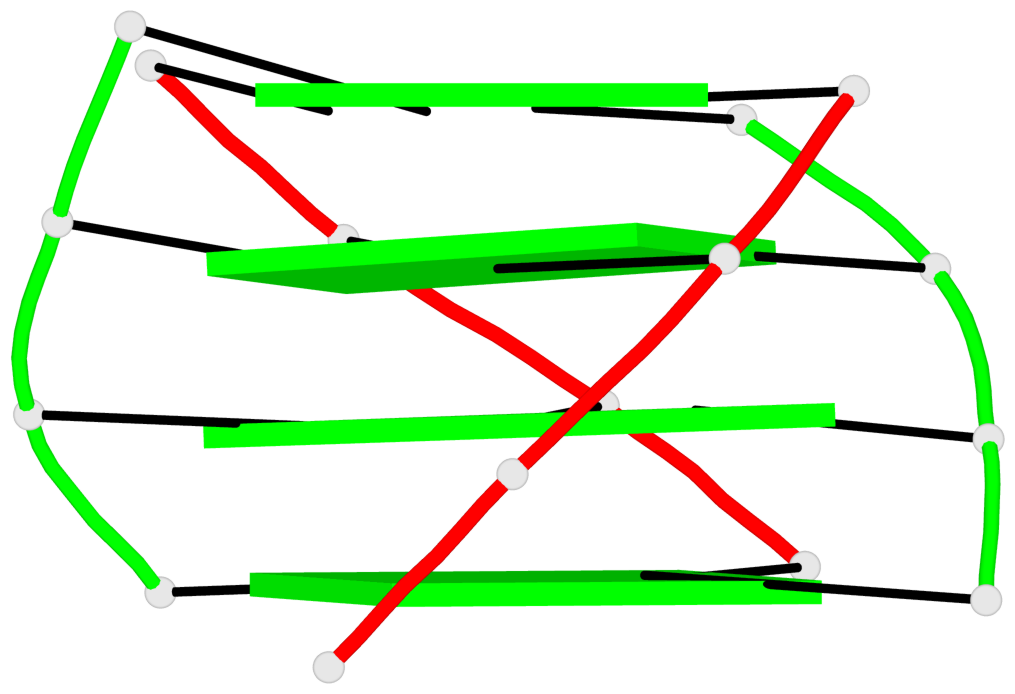
List of 6 G4-helices
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 4 G-tetrad layers, inter-molecular, with 1 stem
Helix#2, 4 G-tetrad layers, inter-molecular, with 1 stem
Helix#3, 4 G-tetrad layers, inter-molecular, with 1 stem
Helix#4, 4 G-tetrad layers, inter-molecular, with 1 stem
Helix#5, 4 G-tetrad layers, inter-molecular, with 1 stem
Helix#6, 4 G-tetrad layers, inter-molecular, with 1 stem
List of 6 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.