Detailed DSSR results for the G-quadruplex: PDB entry 2hbn
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 2hbn
- Class
- DNA
- Method
- X-ray (1.55 Å)
- Summary
- Crystallization of the tl+-form of the oxytricha nova g-quadruplex
- Reference
- Gill ML, Strobel SA, Loria JP (2006): "Crystallization and characterization of the thallium form of the Oxytricha nova G-quadruplex." Nucleic Acids Res., 34, 4506-4514. doi: 10.1093/nar/gkl616.
- Abstract
- The crystal structure of the Tl+ form of the G-quadruplex formed from the Oxytricha nova telomere sequence, d(G4T4G4), has been solved to 1.55 A. This G-quadruplex contains five Tl+ ions, three of which are interspersed between adjacent G-quartet planes and one in each of the two thymine loops. The structure displays a high degree of similarity to the K+ crystal structure [Haider et al. (2002), J. Mol. Biol., 320, 189-200], including the number and location of the monovalent cation binding sites. The highly isomorphic nature of the two structures, which contain such a large number of monovalent binding sites (relative to nucleic acid content), verifies the ability of Tl+ to mimic K+ in nucleic acids. Information from this report confirms and extends the assignment of 205Tl resonances from a previous report [Gill et al. (2005), J. Am. Chem. Soc., 127, 16 723-16 732] where 205Tl NMR was used to study monovalent cation binding to this G-quadruplex. The assignment of these resonances provides evidence for the occurrence of conformational dynamics in the thymine loop region that is in slow exchange on the 205Tl timescale.
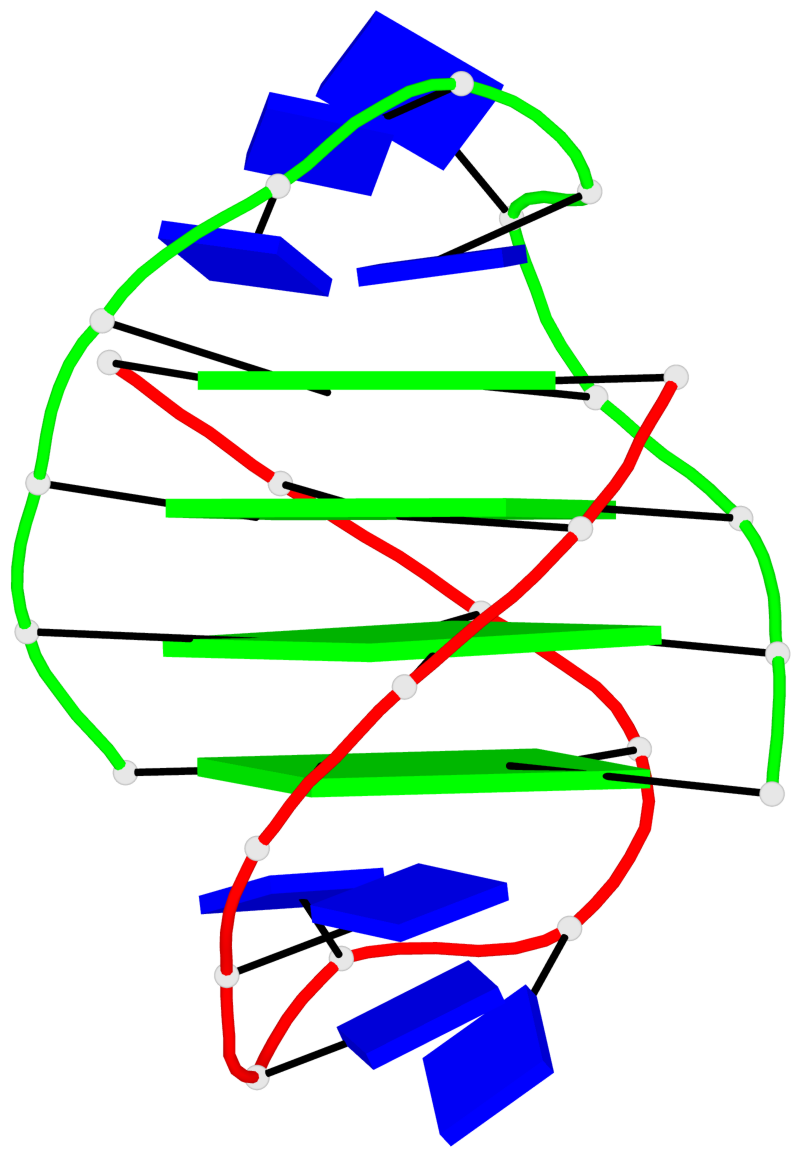
- G4 notes
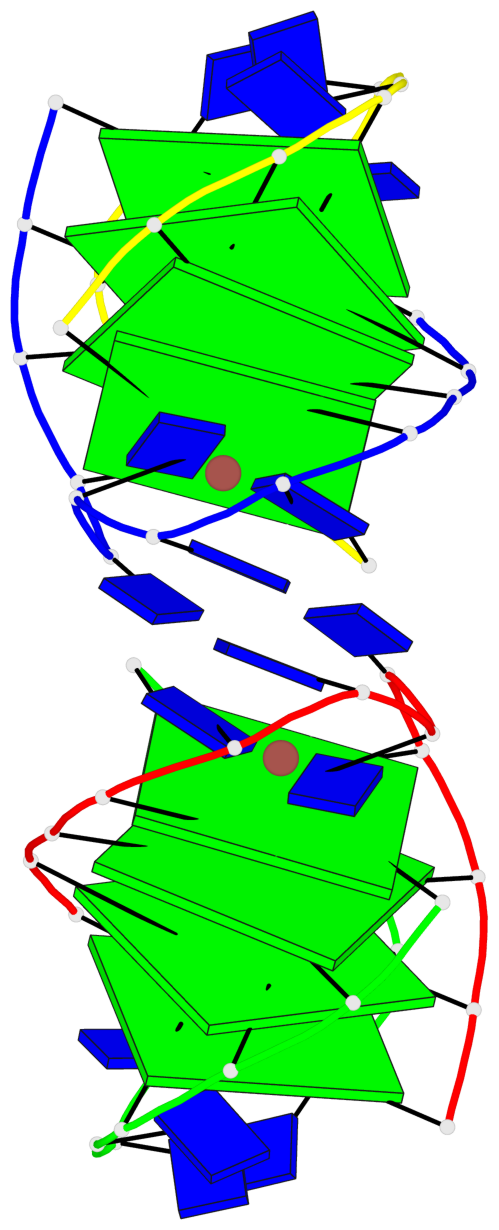
- 8 G-tetrads, 2 G4 helices, 2 G4 stems, (2+2), UDDU
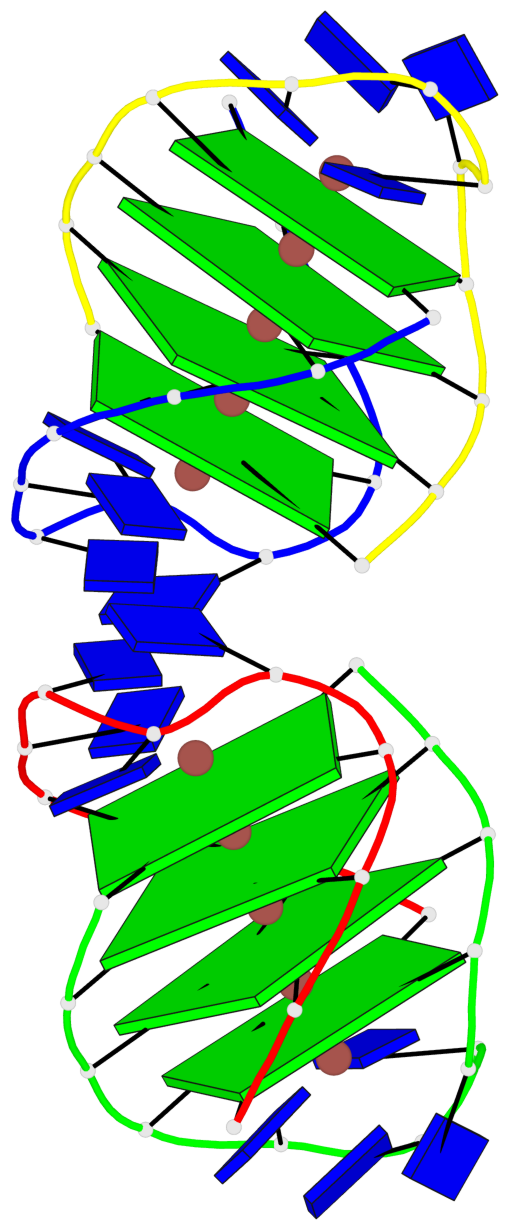
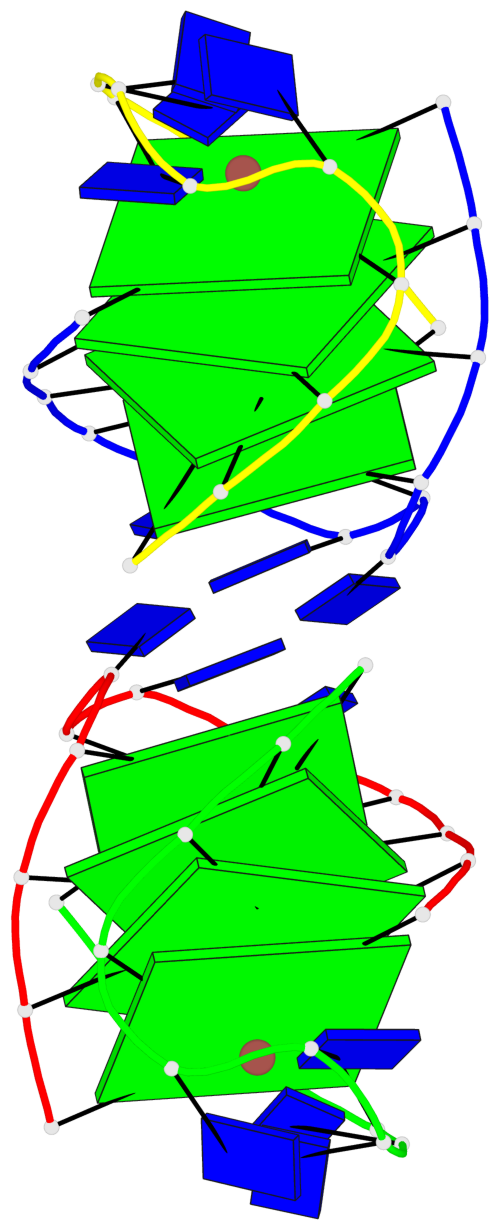
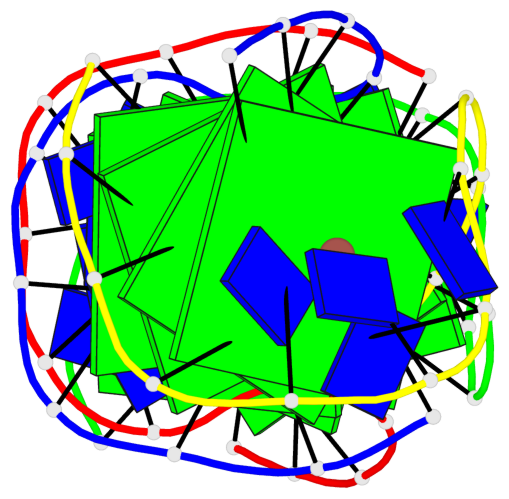
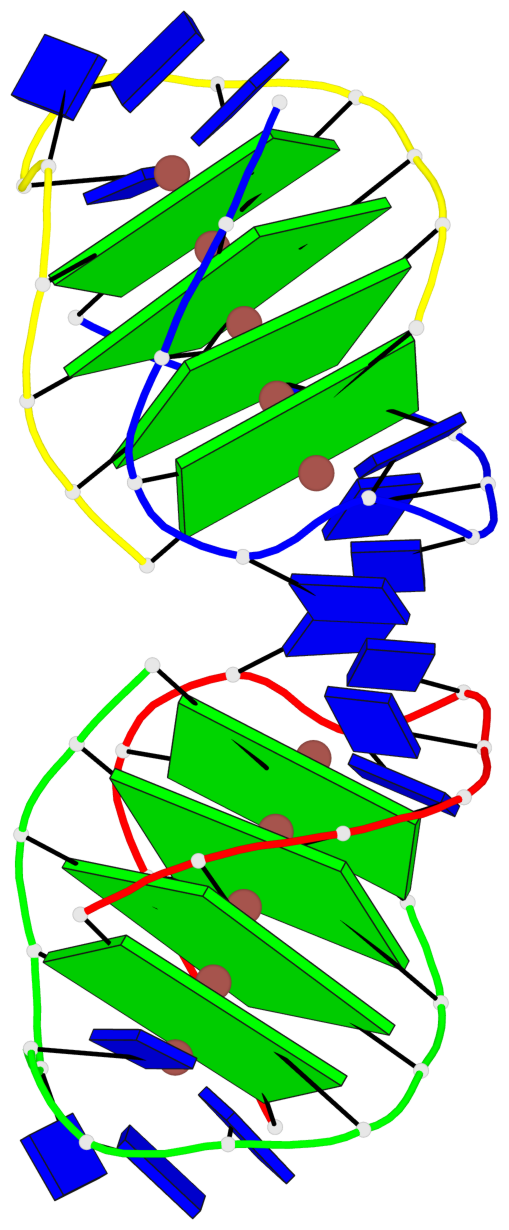
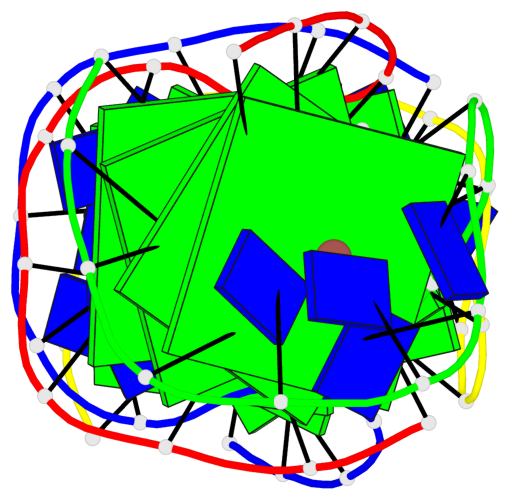
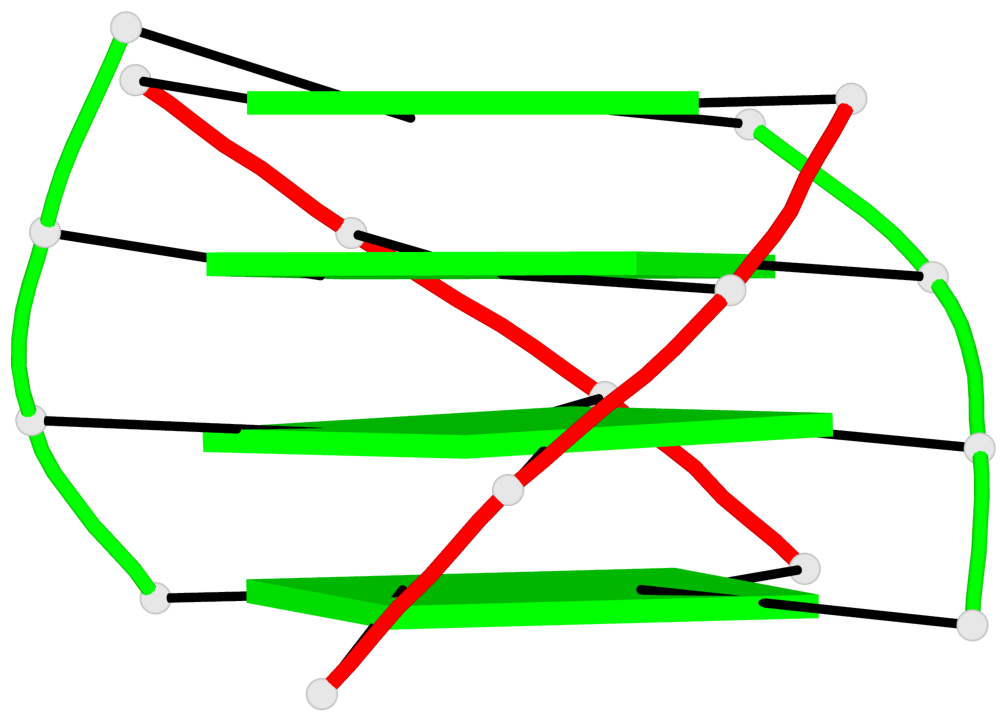
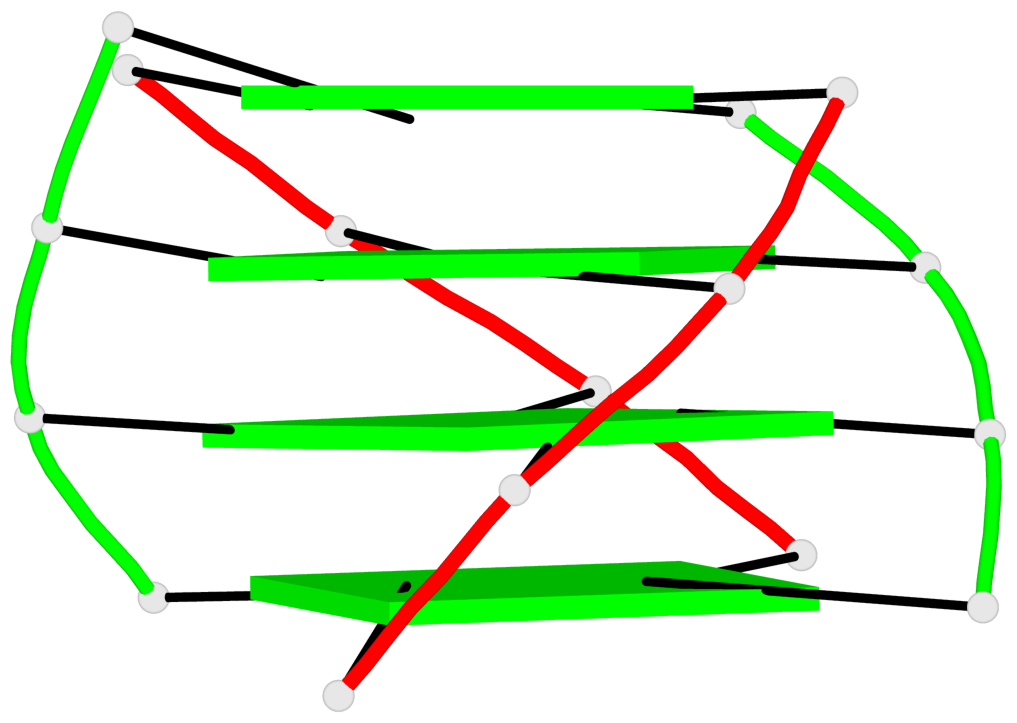
Base-block schematics in six views
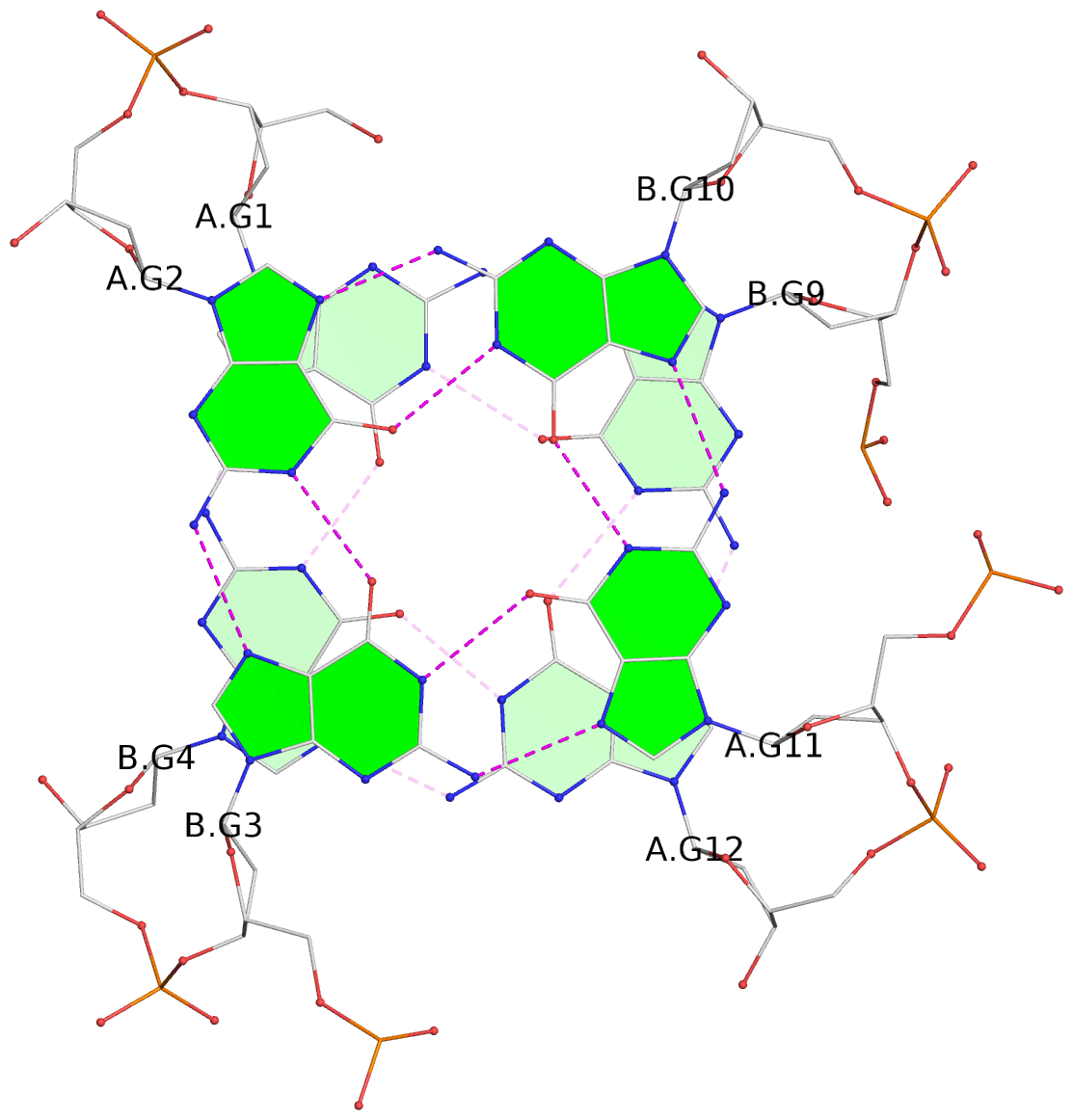
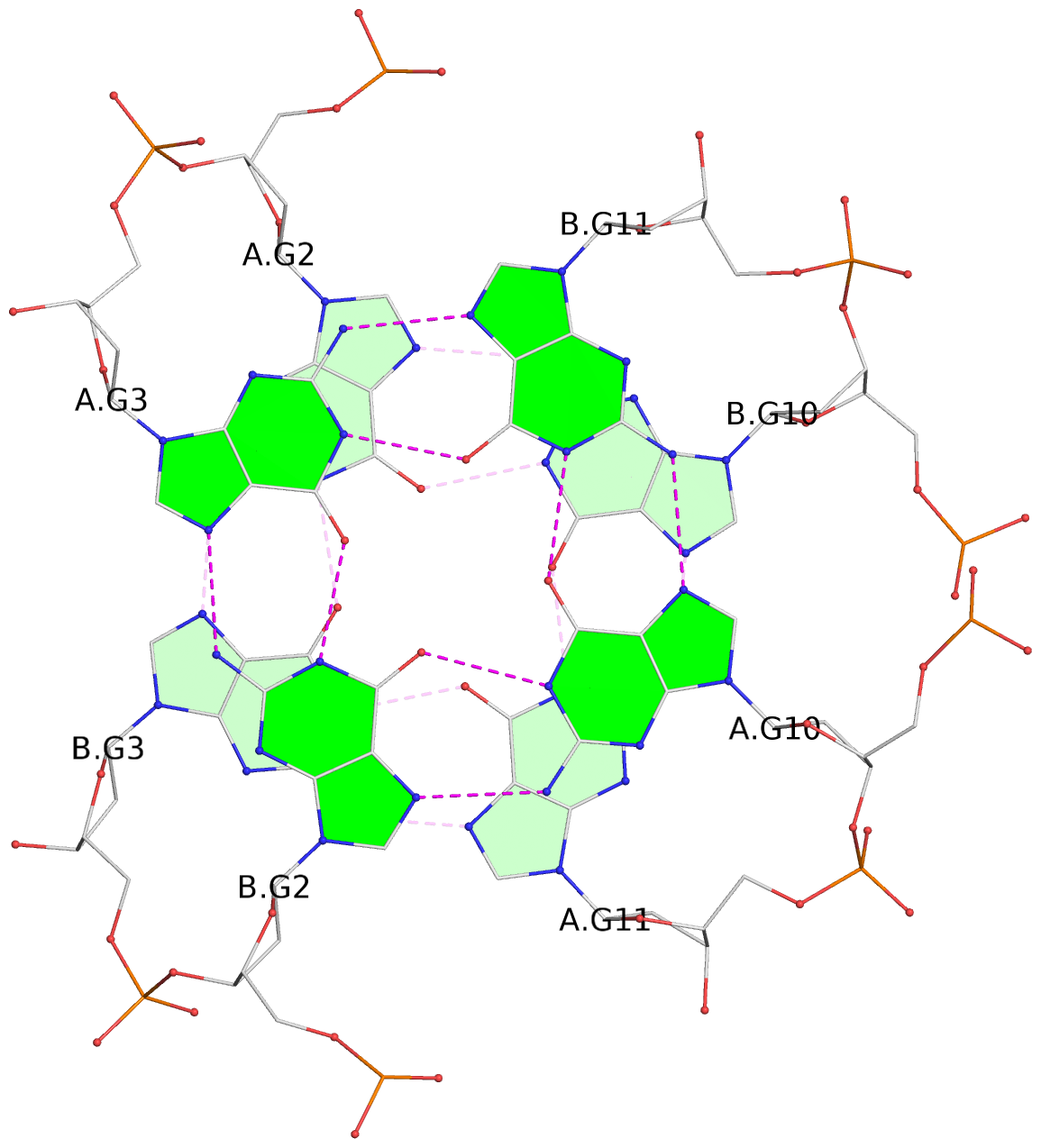
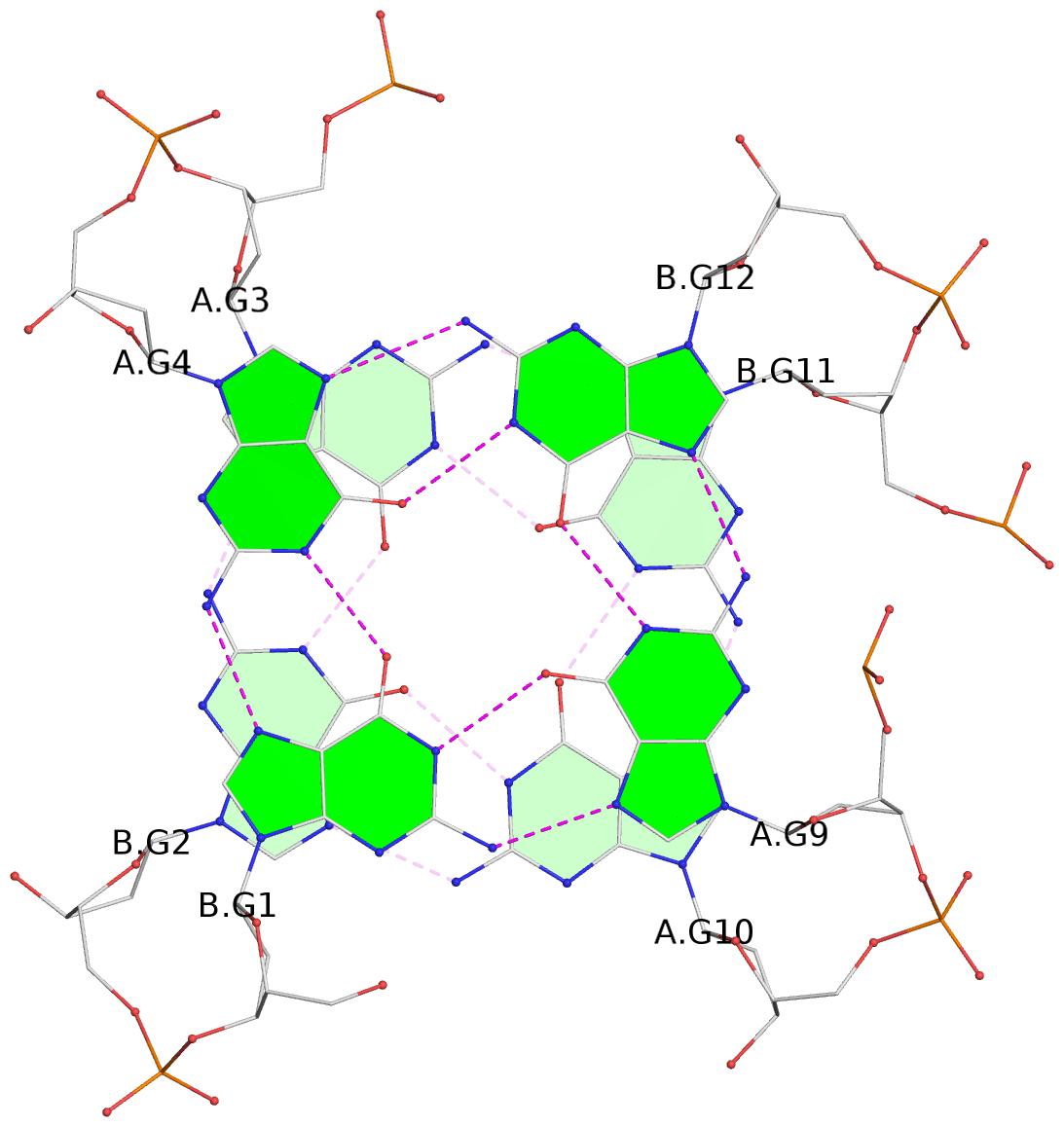
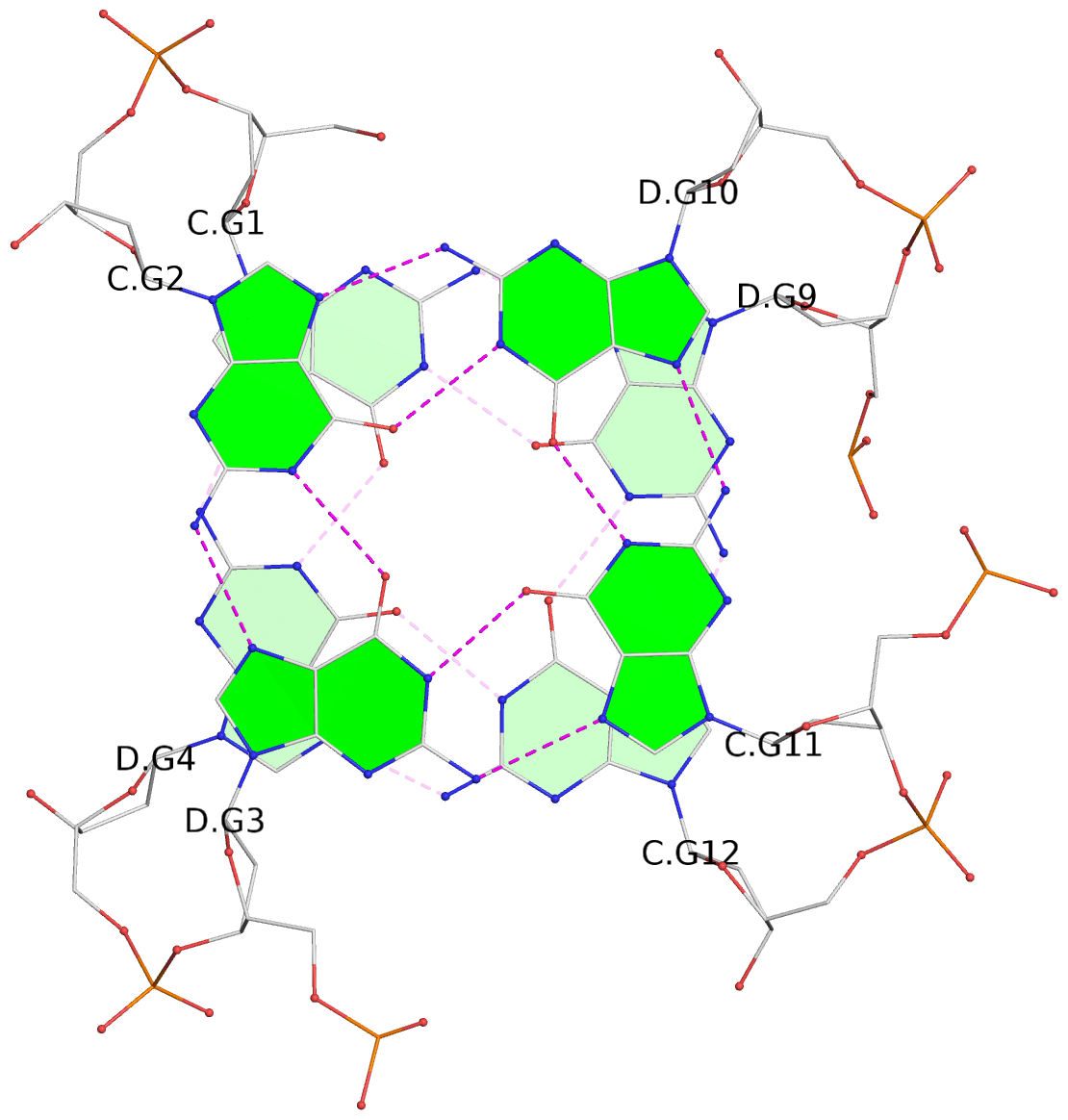
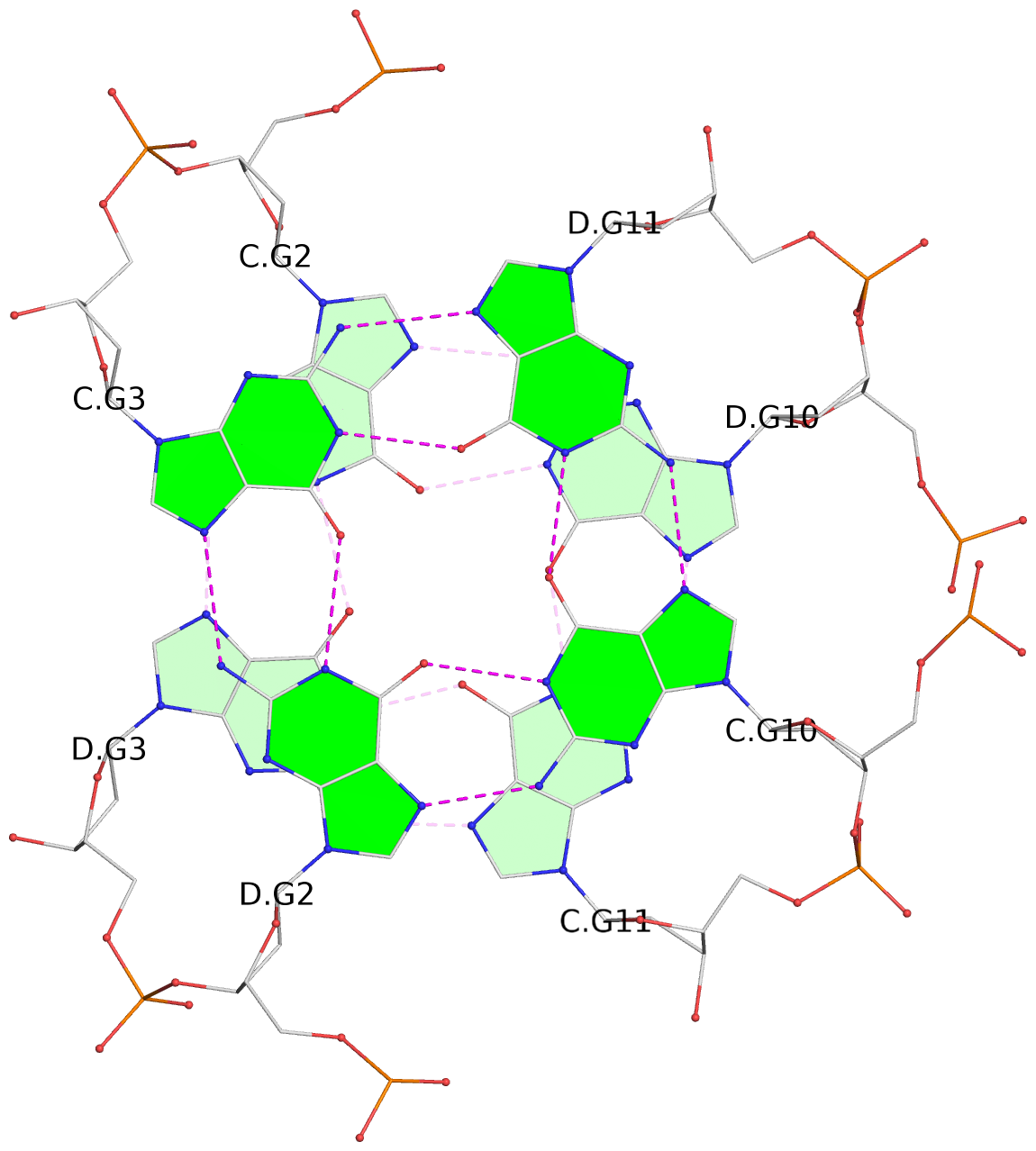
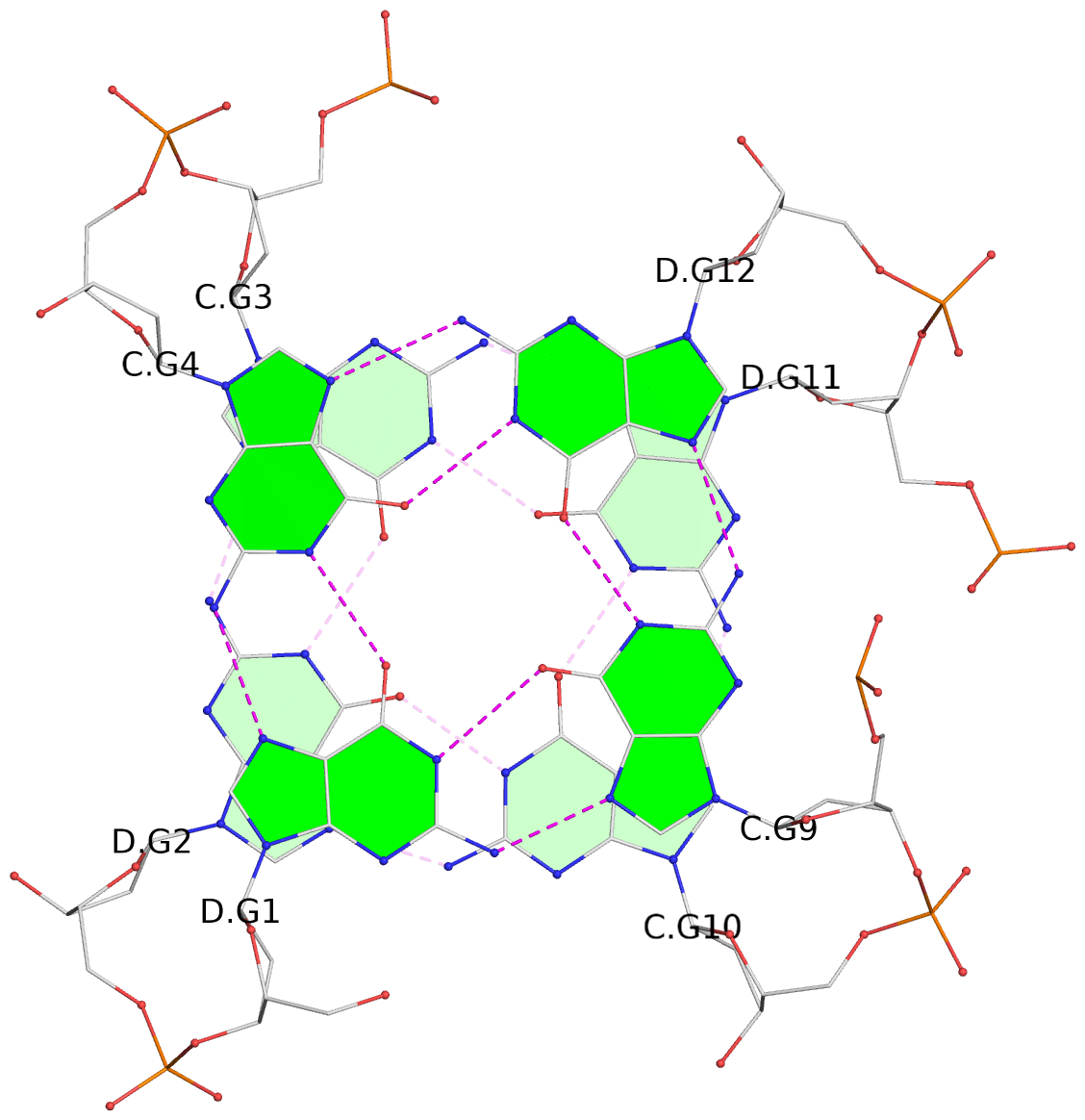
List of 8 G-tetrads
1 glyco-bond=s--s sugar=---- groove=w-n- planarity=0.205 type=other nts=4 GGGG A.DG1,B.DG4,A.DG12,B.DG9 2 glyco-bond=-ss- sugar=---- groove=w-n- planarity=0.157 type=planar nts=4 GGGG A.DG2,B.DG3,A.DG11,B.DG10 3 glyco-bond=s--s sugar=---- groove=w-n- planarity=0.153 type=planar nts=4 GGGG A.DG3,B.DG2,A.DG10,B.DG11 4 glyco-bond=-ss- sugar=---- groove=w-n- planarity=0.233 type=other nts=4 GGGG A.DG4,B.DG1,A.DG9,B.DG12 5 glyco-bond=s--s sugar=---- groove=w-n- planarity=0.240 type=other nts=4 GGGG C.DG1,D.DG4,C.DG12,D.DG9 6 glyco-bond=-ss- sugar=---- groove=w-n- planarity=0.170 type=other nts=4 GGGG C.DG2,D.DG3,C.DG11,D.DG10 7 glyco-bond=s--s sugar=---- groove=w-n- planarity=0.163 type=other nts=4 GGGG C.DG3,D.DG2,C.DG10,D.DG11 8 glyco-bond=-ss- sugar=---- groove=w-n- planarity=0.239 type=other nts=4 GGGG C.DG4,D.DG1,C.DG9,D.DG12
List of 2 G4-helices
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 4 G-tetrad layers, inter-molecular, with 1 stem
Helix#2, 4 G-tetrad layers, inter-molecular, with 1 stem
List of 2 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.