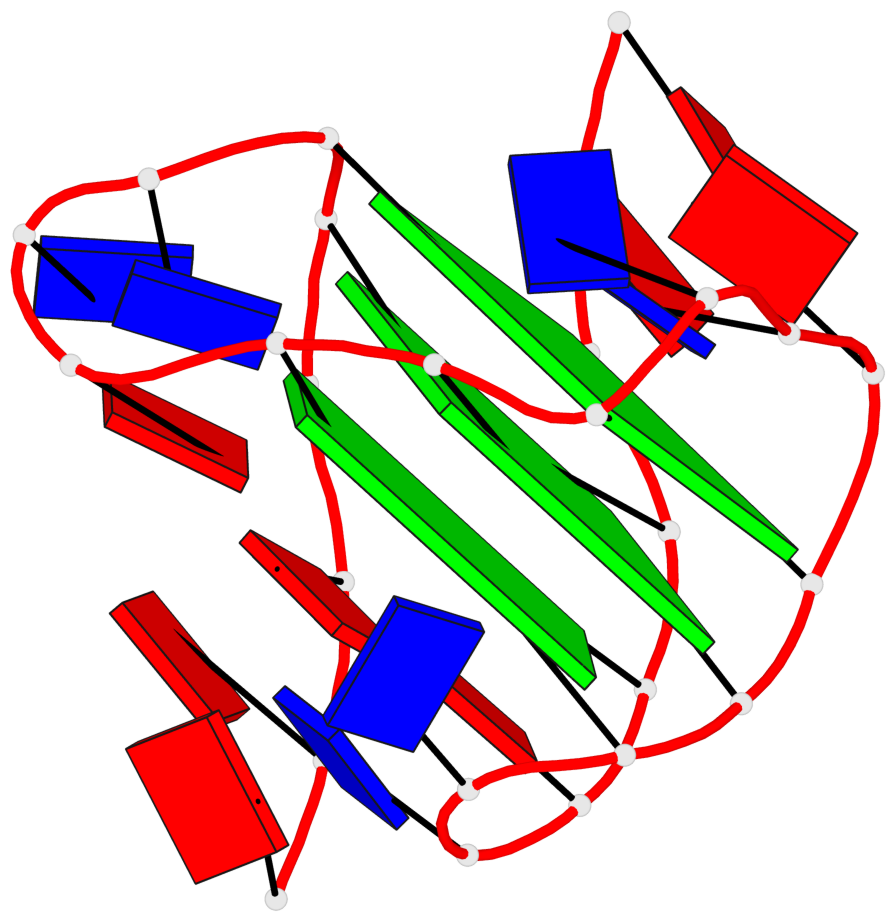
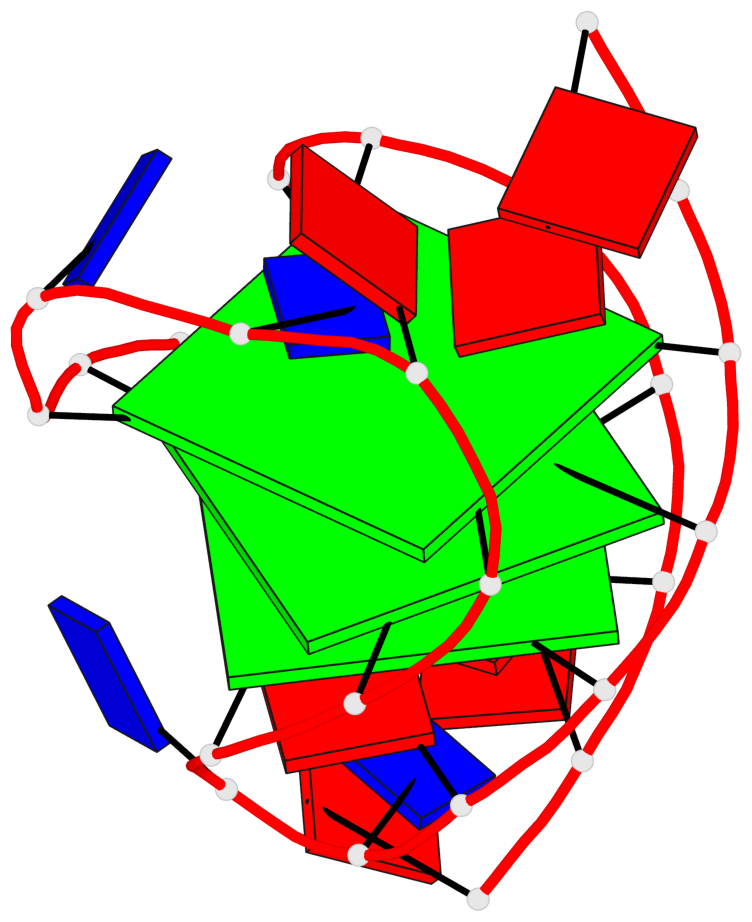
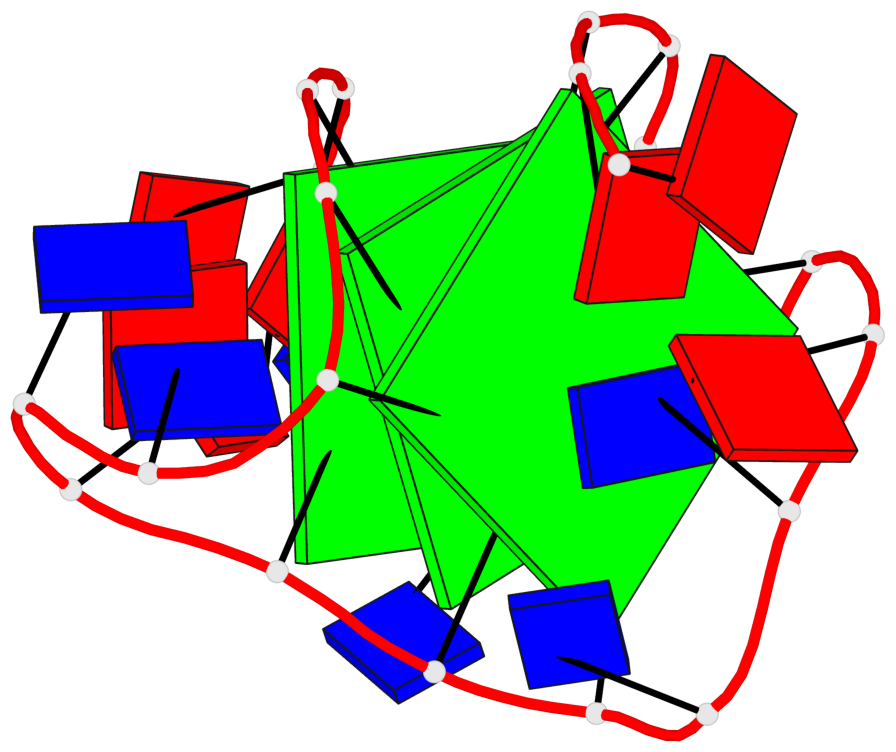
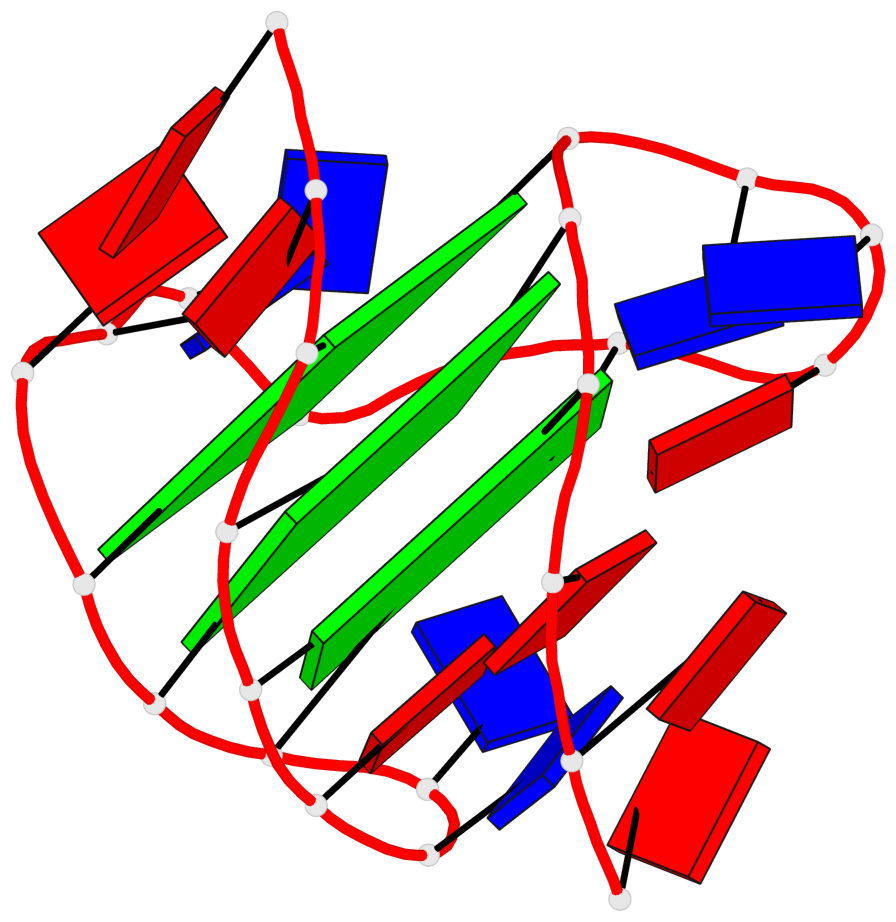
Detailed DSSR results for the G-quadruplex: PDB entry 2hy9
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 2hy9
- Class
- DNA
- Method
- NMR
- Summary
- Human telomere DNA quadruplex structure in k+ solution hybrid-1 form
- Reference
- Dai J, Punchihewa C, Ambrus A, Chen D, Jones RA, Yang D (2007): "Structure of the intramolecular human telomeric G-quadruplex in potassium solution: a novel adenine triple formation." Nucleic Acids Res., 35, 2440-2450. doi: 10.1093/nar/gkm009.
- Abstract
- We report the NMR solution structure of the intramolecular G-quadruplex formed in human telomeric DNA in K(+). The hybrid-type telomeric G-quadruplex consists of three G-tetrads linked with mixed parallel-antiparallel G-strands, with the bottom two G-tetrads having the same G-arrangement (anti:anti:syn:anti) and the top G-tetrad having the reversed G-arrangement (syn:syn:anti:syn). The three TTA loop segments adopt different conformations, with the first TTA assuming a double-chain-reversal loop conformation, and the second and third TTA assuming lateral loop conformations. The NMR structure is very well defined, including the three TTA loops and the two flanking sequences at 5'- and 3'-ends. Our study indicates that the three loop regions interact with the core G-tetrads in a specific way that defines and stabilizes the unique human telomeric G-quadruplex structure in K(+). Significantly, a novel adenine triple platform is formed with three naturally occurring adenine residues, A21, A3 and A9, capping the top tetrad of the hybrid-type telomeric G-quadruplex. This adenine triple is likely to play an important role in the formation of a stable human telomeric G-quadruplex structure in K(+). The unique human telomeric G-quadruplex structure formed in K(+) suggests that it can be specifically targeted for anticancer drug design.
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(-P-Lw-Ln), hybrid-1(3+1), UUDU
Base-block schematics in six views
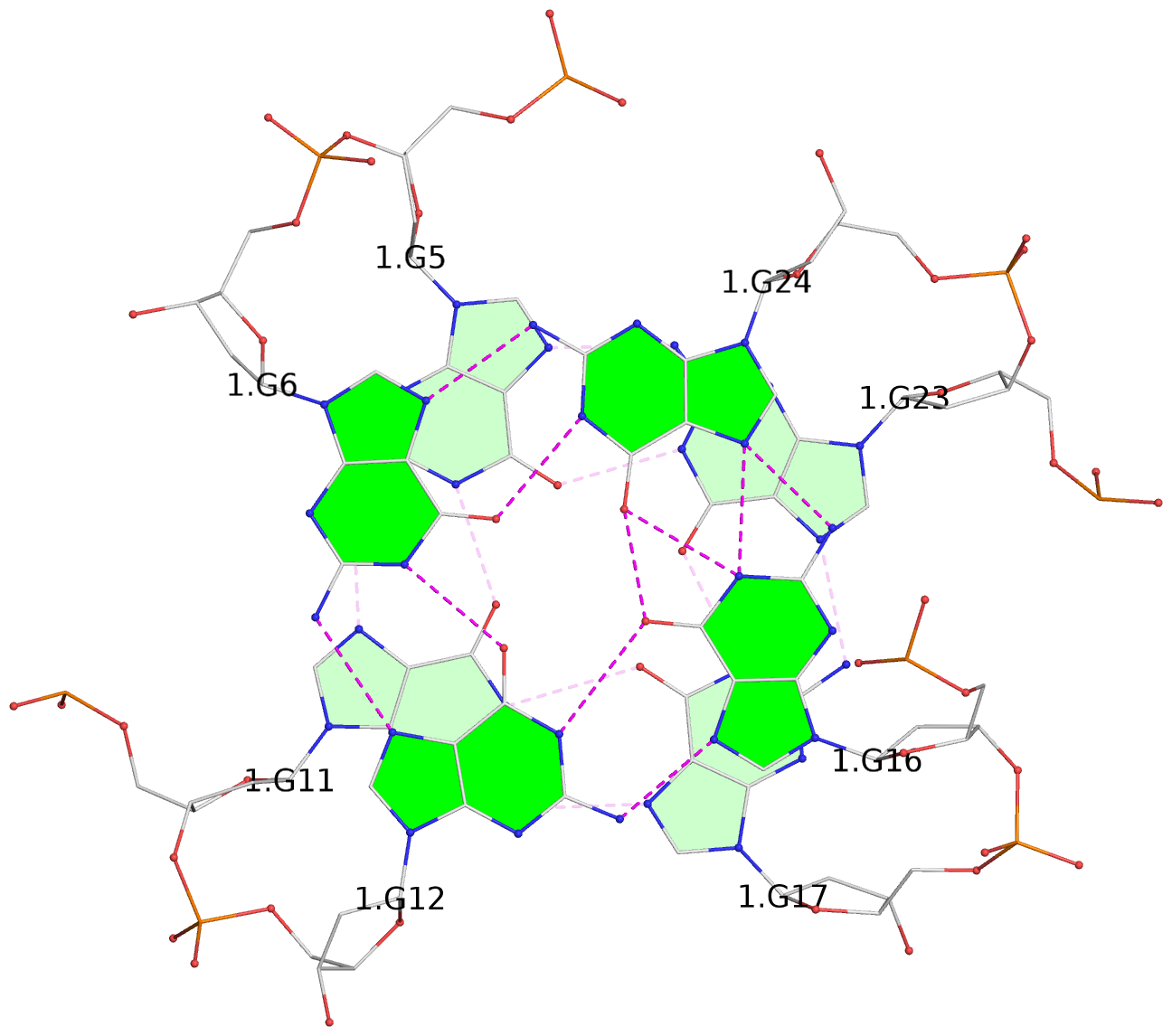
List of 3 G-tetrads
1 glyco-bond=ss-s sugar=-... groove=-wn- planarity=0.077 type=planar nts=4 GGGG 1.DG4,1.DG10,1.DG18,1.DG22 2 glyco-bond=--s- sugar=.... groove=-wn- planarity=0.066 type=planar nts=4 GGGG 1.DG5,1.DG11,1.DG17,1.DG23 3 glyco-bond=--s- sugar=---- groove=-wn- planarity=0.076 type=planar nts=4 GGGG 1.DG6,1.DG12,1.DG16,1.DG24
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.