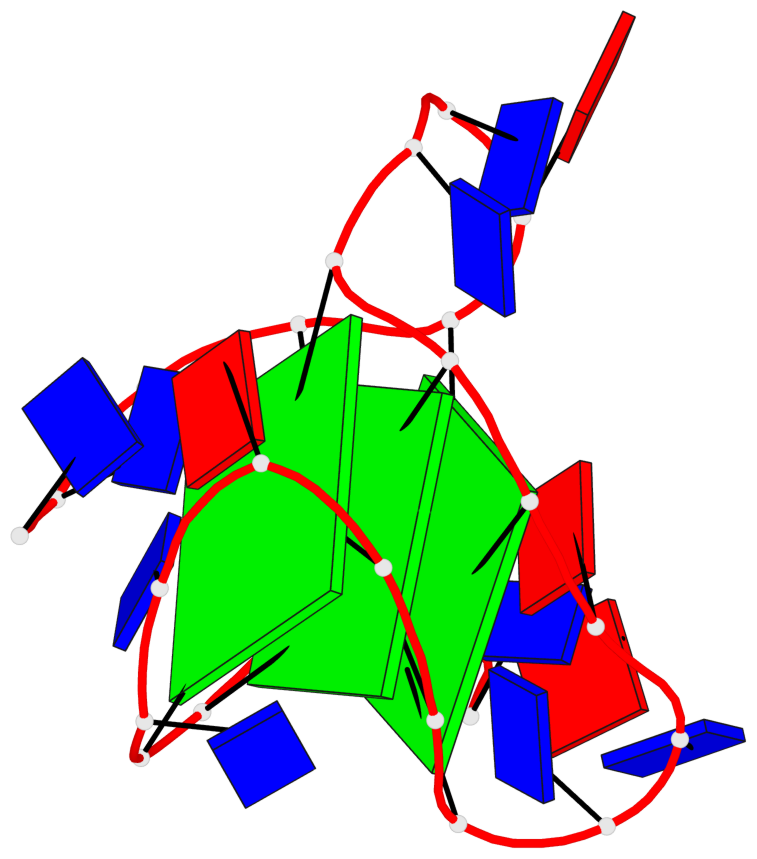
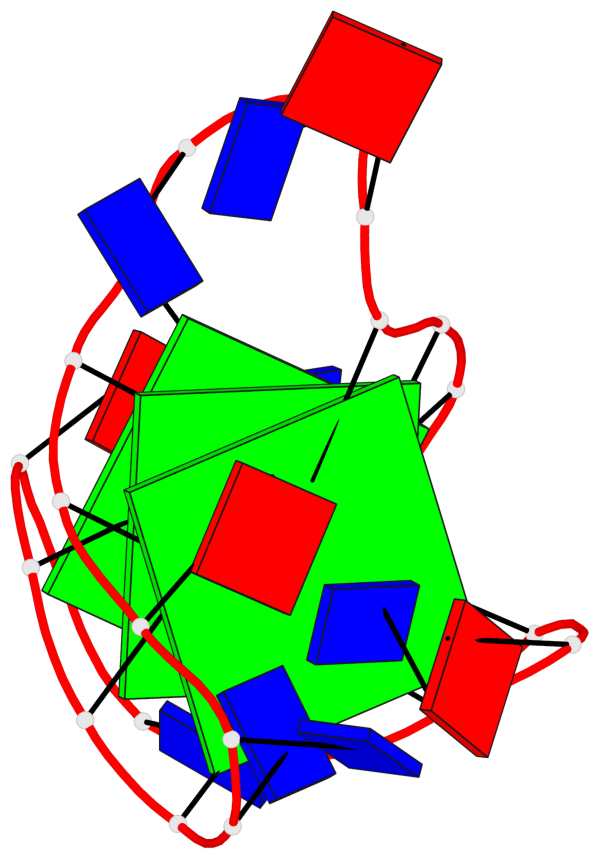
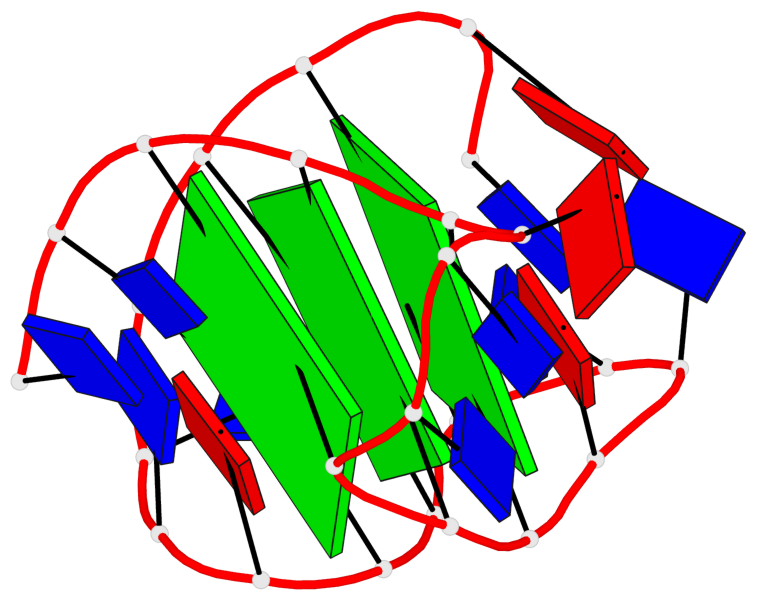
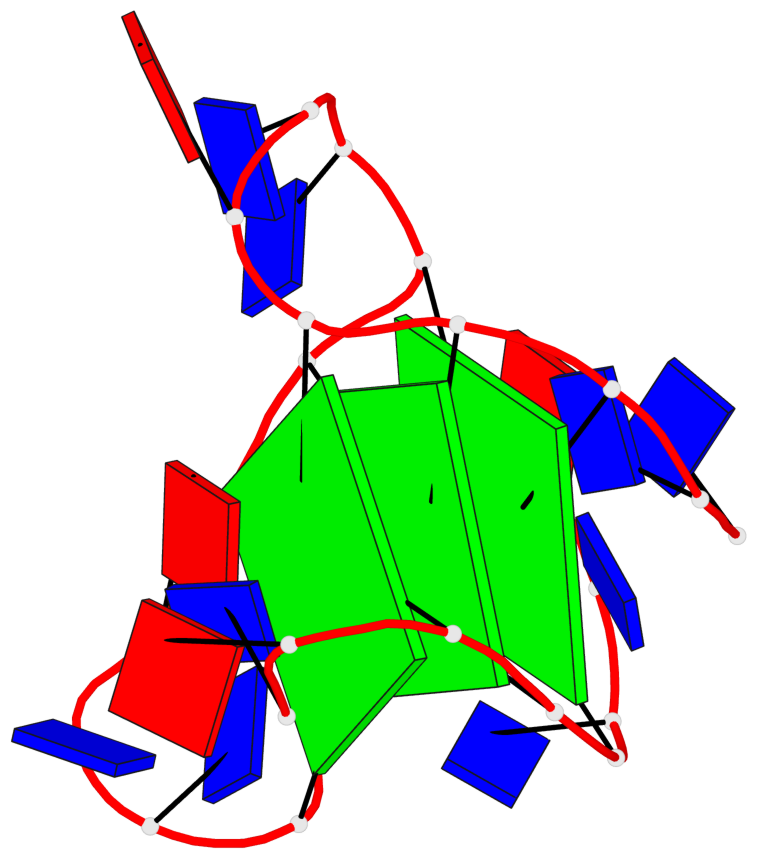
Detailed DSSR results for the G-quadruplex: PDB entry 2jsl
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 2jsl
- Class
- DNA
- Method
- NMR
- Summary
- Monomeric human telomere DNA tetraplex with 3+1 strand fold topology, two edgewise loops and double-chain reversal loop, form 2 natural, NMR, 10 structures
- Reference
- Phan AT, Kuryavyi V, Luu KN, Patel DJ (2007): "Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution." Nucleic Acids Res., 35, 6517-6525. doi: 10.1093/nar/gkm706.
- Abstract
- Intramolecular G-quadruplexes formed by human telomere sequences are attractive anticancer targets. Recently, four-repeat human telomere sequences have been shown to form two different intramolecular (3 + 1) G-quadruplexes in K(+) solution (Form 1 and Form 2). Here we report on the solution structures of both Form 1 and Form 2 adopted by natural human telomere sequences. Both structures contain the (3 + 1) G-tetrad core with one double-chain-reversal and two edgewise loops, but differ in the successive order of loop arrangements within the G-quadruplex scaffold. Our results provide the structural details at the two ends of the G-tetrad core in the context of natural sequences and information on different loop conformations. This structural information might be important for our understanding of telomere G-quadruplex structures and for anticancer drug design targeted to such scaffolds.
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(-Lw-Ln-P), hybrid-2(3+1), UDUU
Base-block schematics in six views
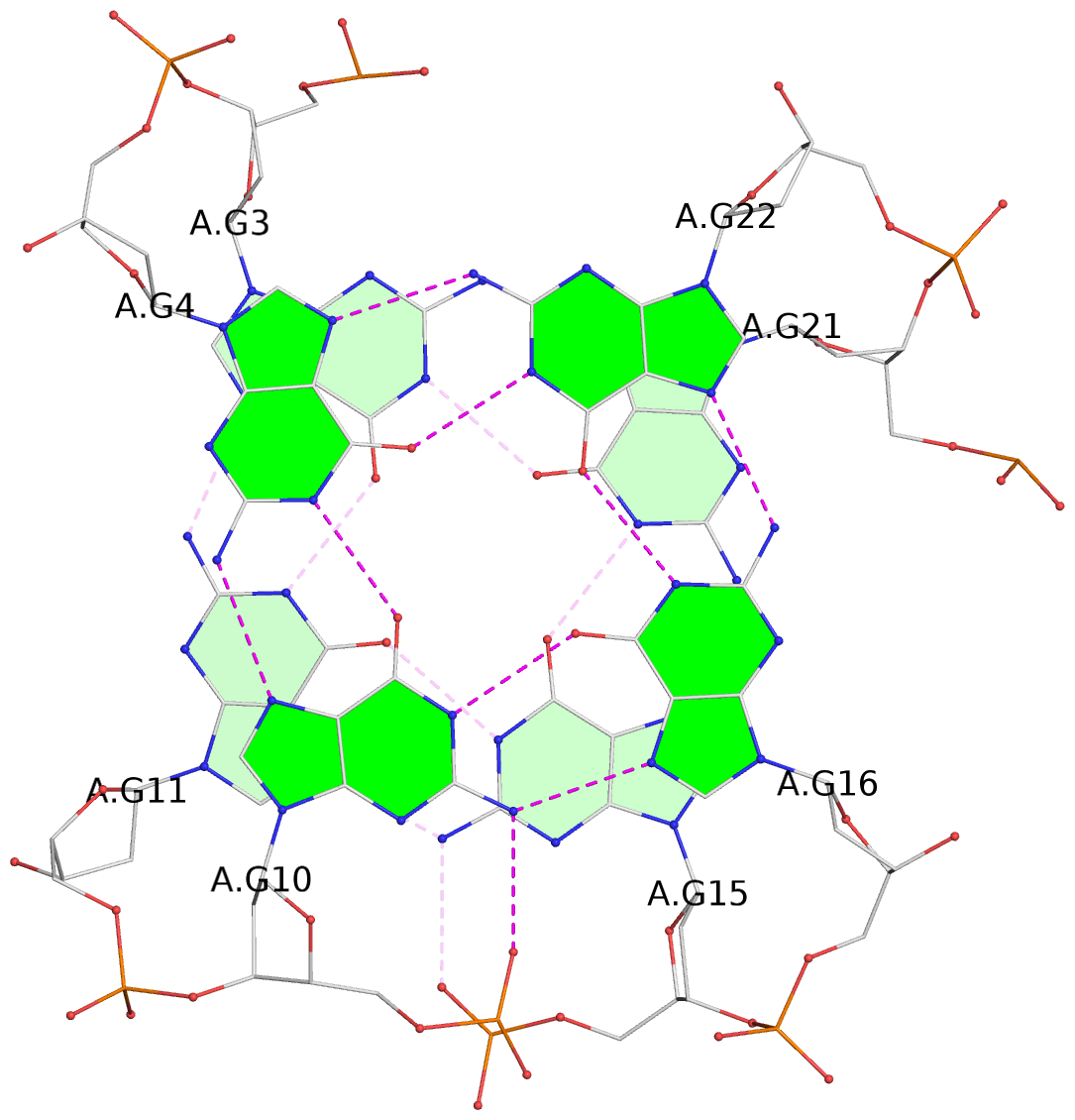
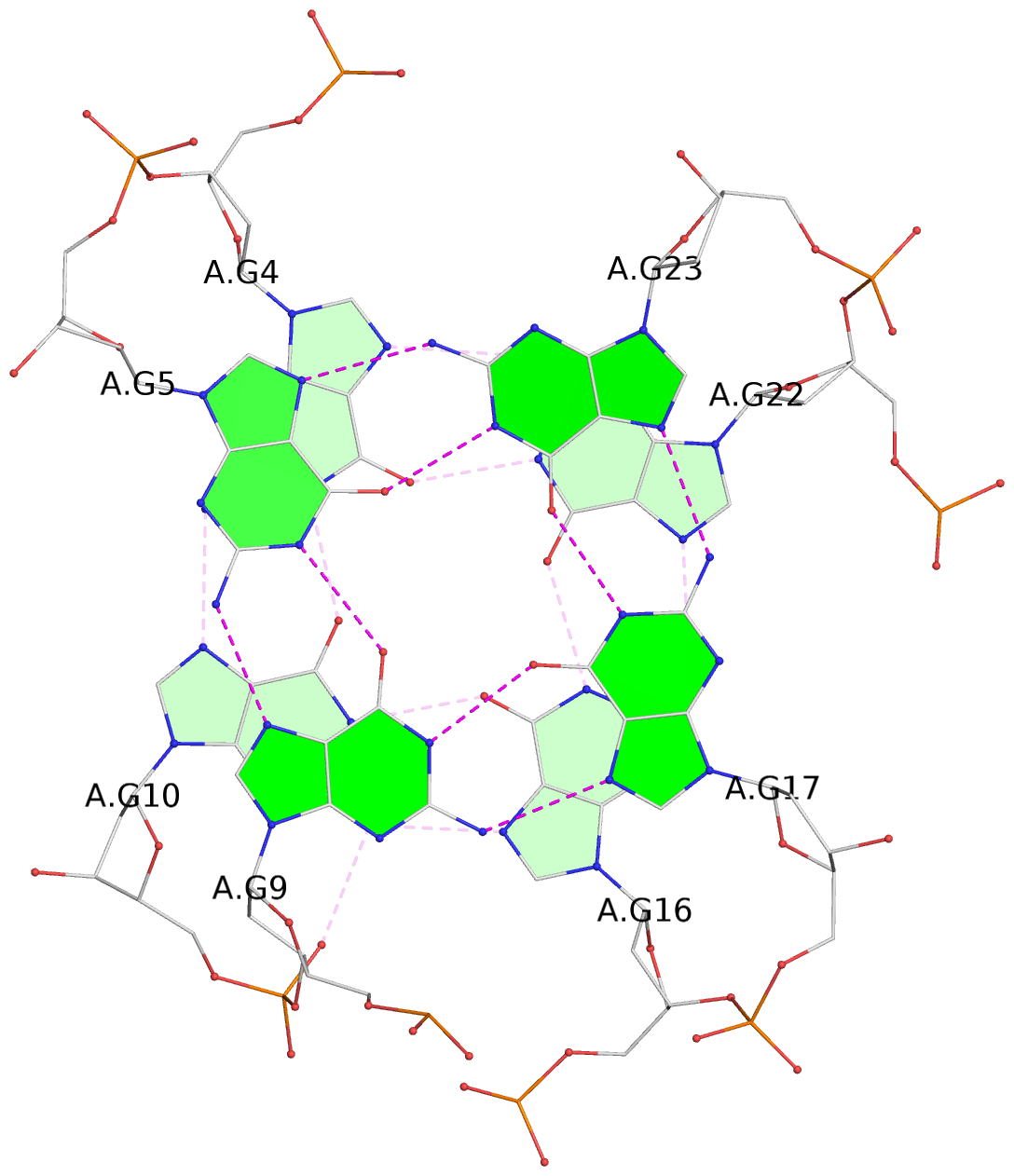
List of 3 G-tetrads
1 glyco-bond=s-ss sugar=---- groove=wn-- planarity=0.195 type=other nts=4 GGGG A.DG3,A.DG11,A.DG15,A.DG21 2 glyco-bond=-s-- sugar=---- groove=wn-- planarity=0.125 type=planar nts=4 GGGG A.DG4,A.DG10,A.DG16,A.DG22 3 glyco-bond=-s-- sugar=-3.- groove=wn-- planarity=0.243 type=other nts=4 GGGG A.DG5,A.DG9,A.DG17,A.DG23
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.