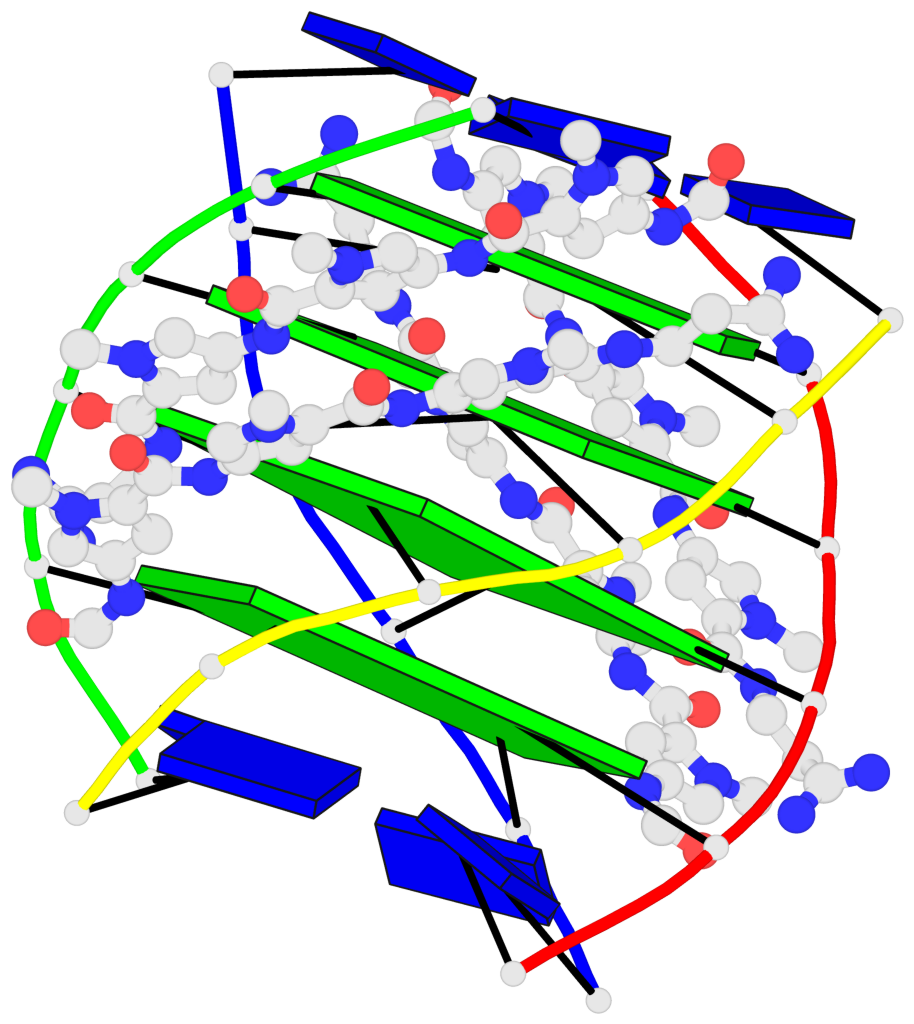
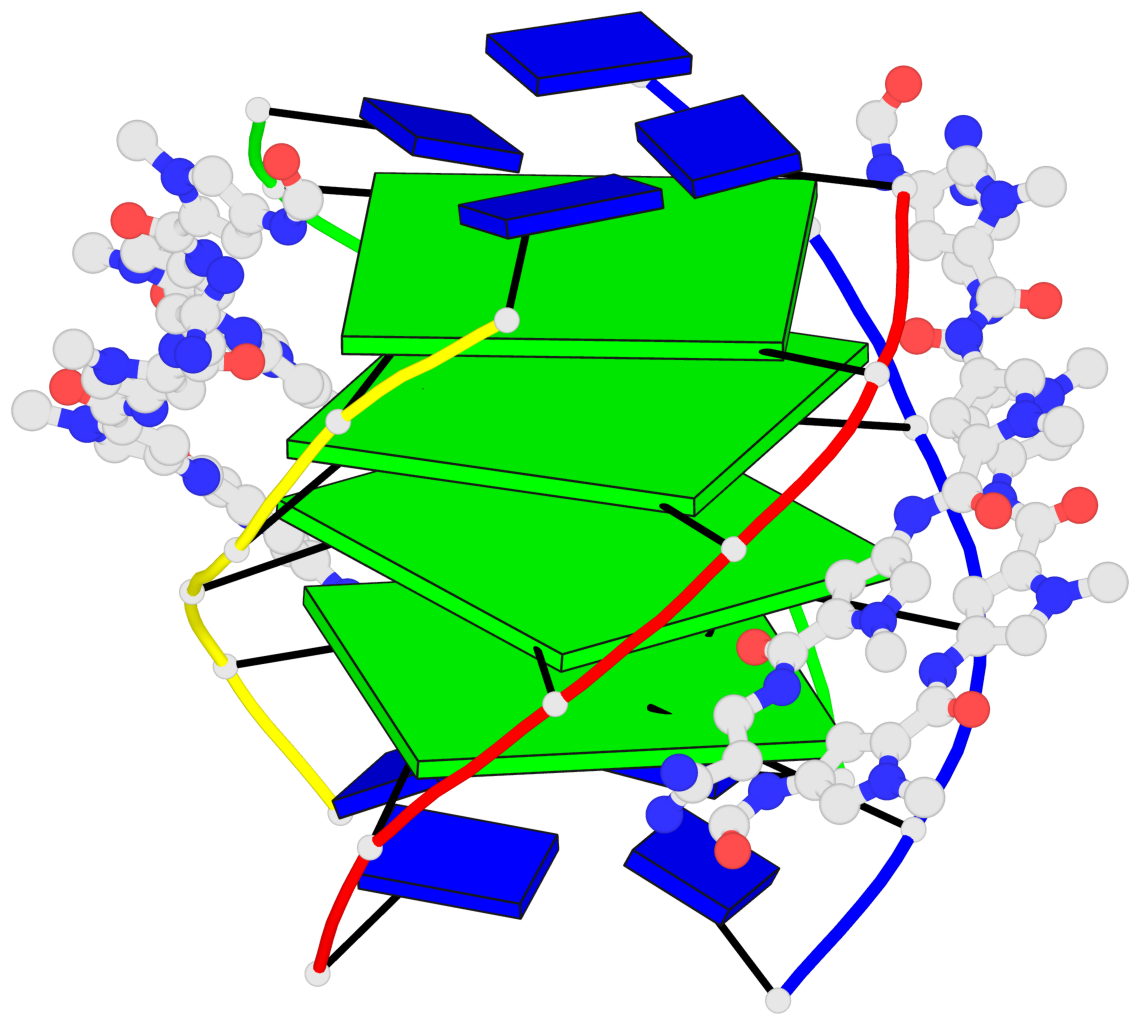
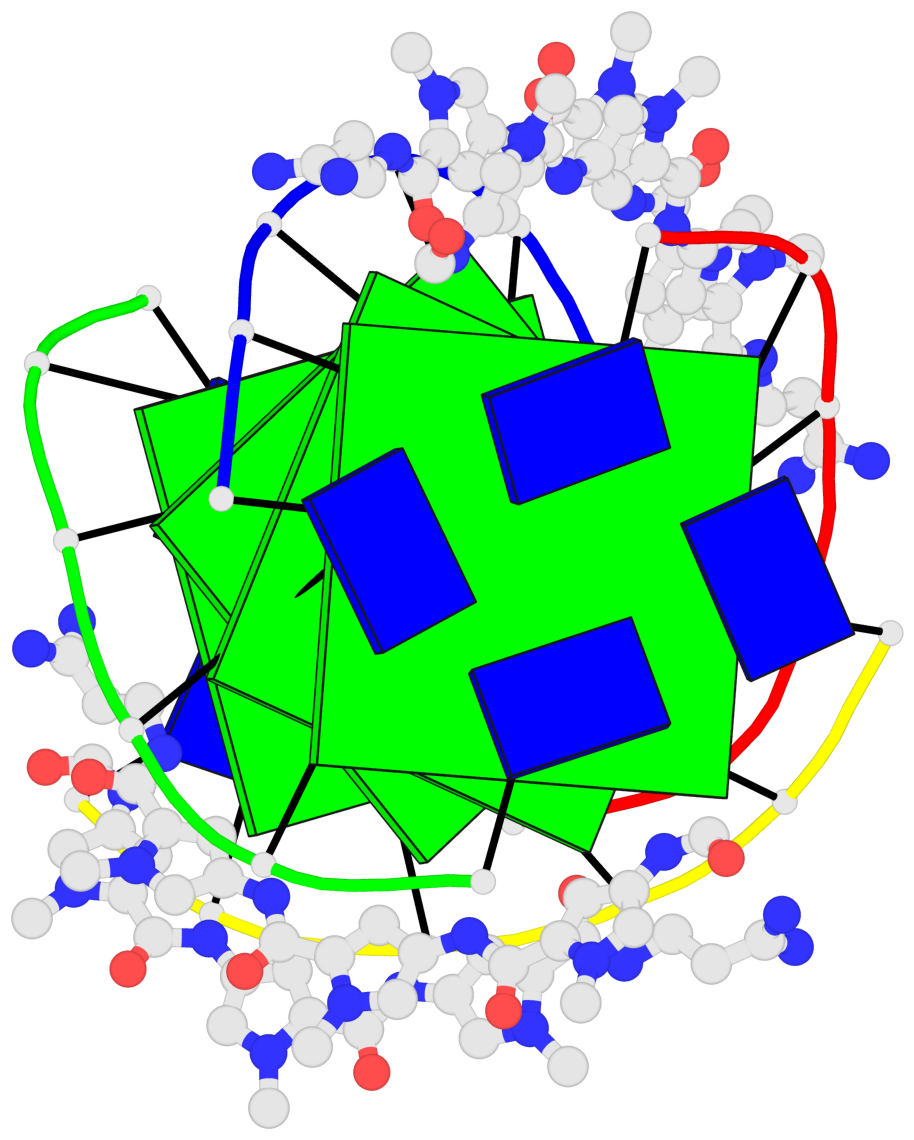
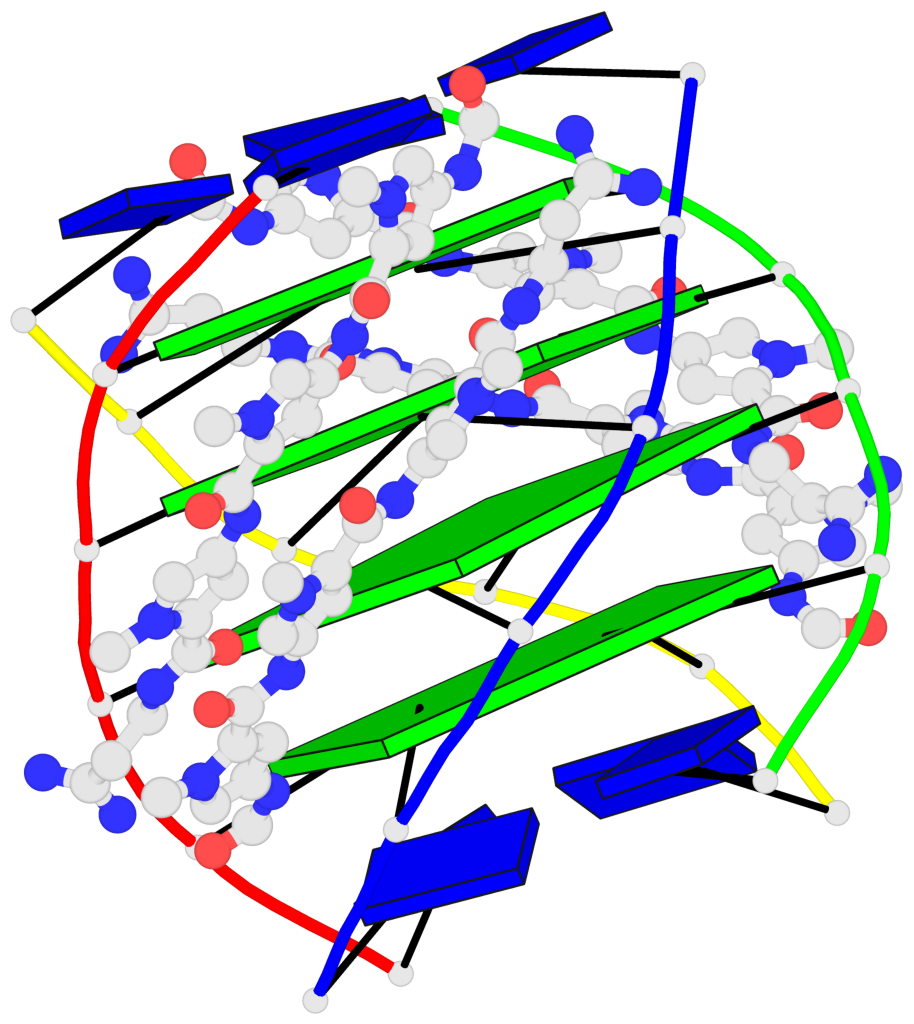
Detailed DSSR results for the G-quadruplex: PDB entry 2jt7
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 2jt7
- Class
- DNA
- Method
- NMR
- Summary
- NMR solution structure of the 4:1 distamycin a-[d(tggggt)]4 complex
- Reference
- Martino L, Virno A, Pagano B, Virgilio A, Di Micco S, Galeone A, Giancola C, Bifulco G, Mayol L, Randazzo A (2007): "Structural and thermodynamic studies of the interaction of distamycin A with the parallel quadruplex structure [d(TGGGGT)]4." J.Am.Chem.Soc., 129, 16048-16056. doi: 10.1021/ja075710k.
- Abstract
- The complex between distamycin A and the parallel DNA quadruplex [d(TGGGGT)]4 has been studied by 1H NMR spectroscopy and isothermal titration calorimetry (ITC). To unambiguously assert that distamycin A interacts with the grooves of the quadruplex [d(TGGGGT)]4, we have analyzed the NMR titration profile of a modified quadruplex, namely [d(TGGMeGGT)]4, and we have applied the recently developed differential frequency-saturation transfer difference (DF-STD) method, for assessing the ligand-DNA binding mode. The three-dimensional structure of the 4:1 distamycin A/[d(TGGGGT)]4 complex has been determined by an in-depth NMR study followed by dynamics and mechanics calculations. All results unequivocally indicate that distamycin molecules interact with [d(TGGGGT)]4 in a 4:1 binding mode, with two antiparallel distamycin dimers that bind simultaneously two opposite grooves of the quadruplex. The affinity between distamycin A and [d(TGGGGT)]4 enhances ( approximately 10-fold) when the ratio of distamycin A to the quadruplex is increased. In this paper we report the first three-dimensional structure of a groove-binder molecule complexed to a DNA quadruplex structure.
- G4 notes
- 4 G-tetrads, 1 G4 helix, 1 G4 stem, parallel(4+0), UUUU
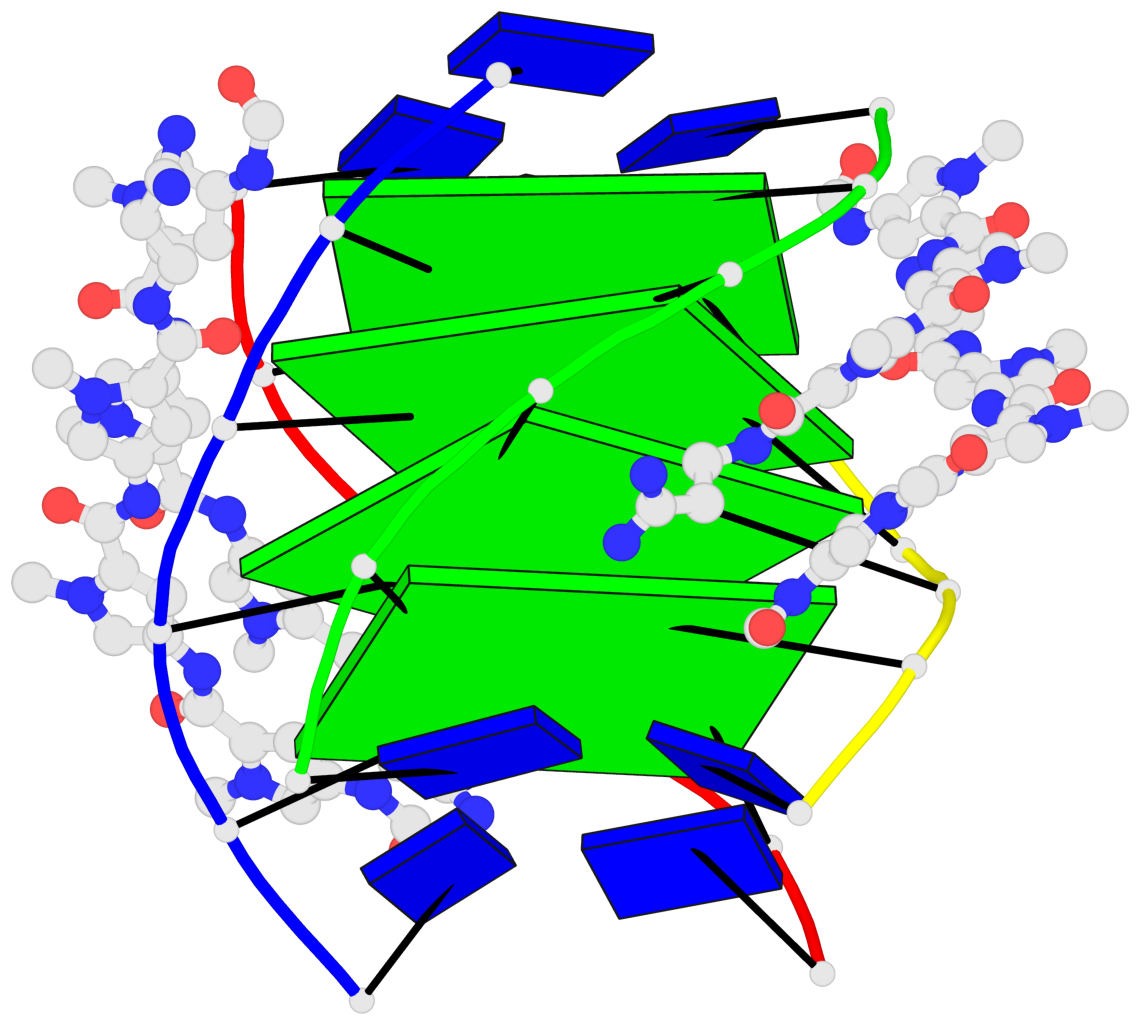
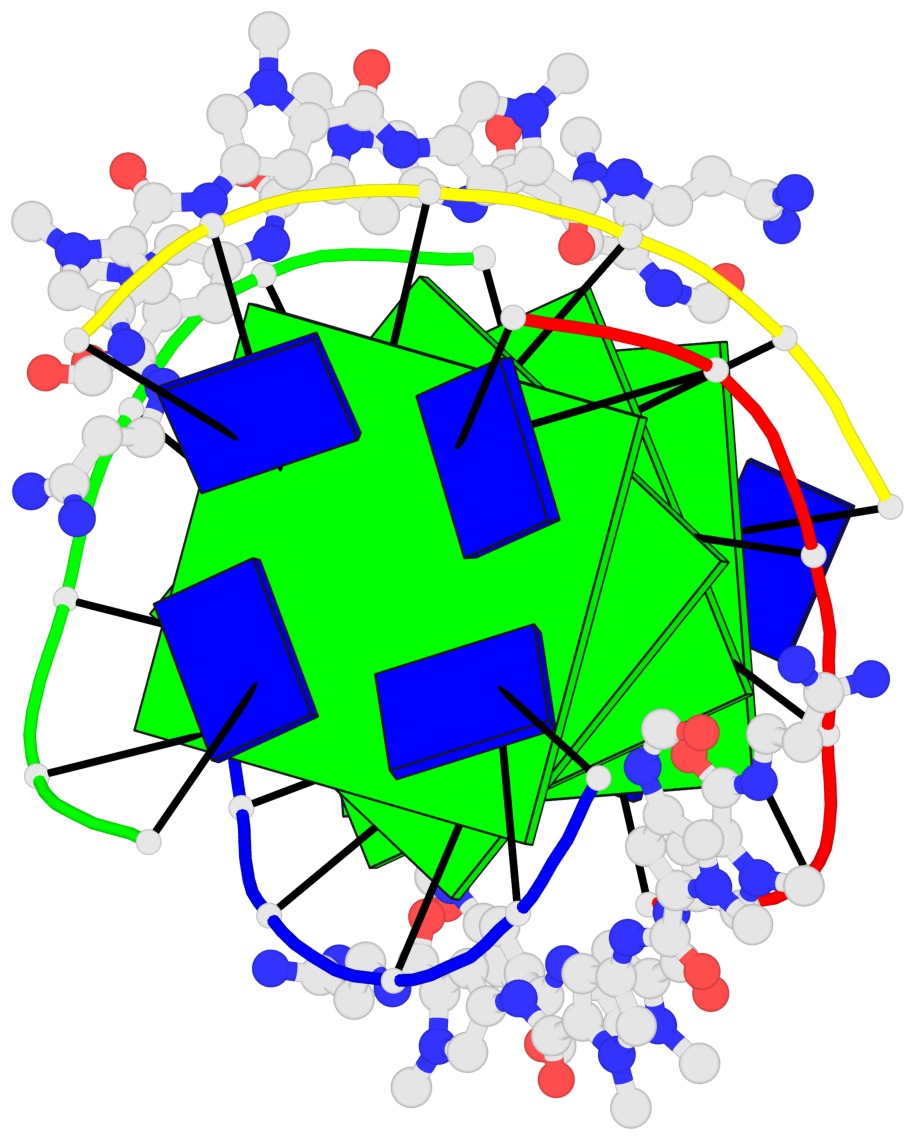
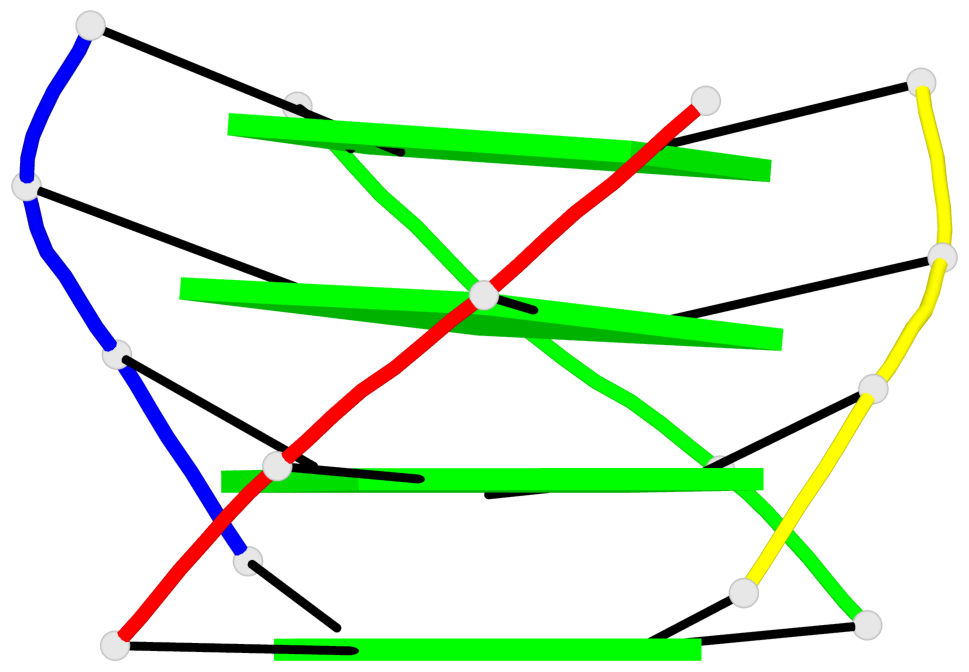
Base-block schematics in six views
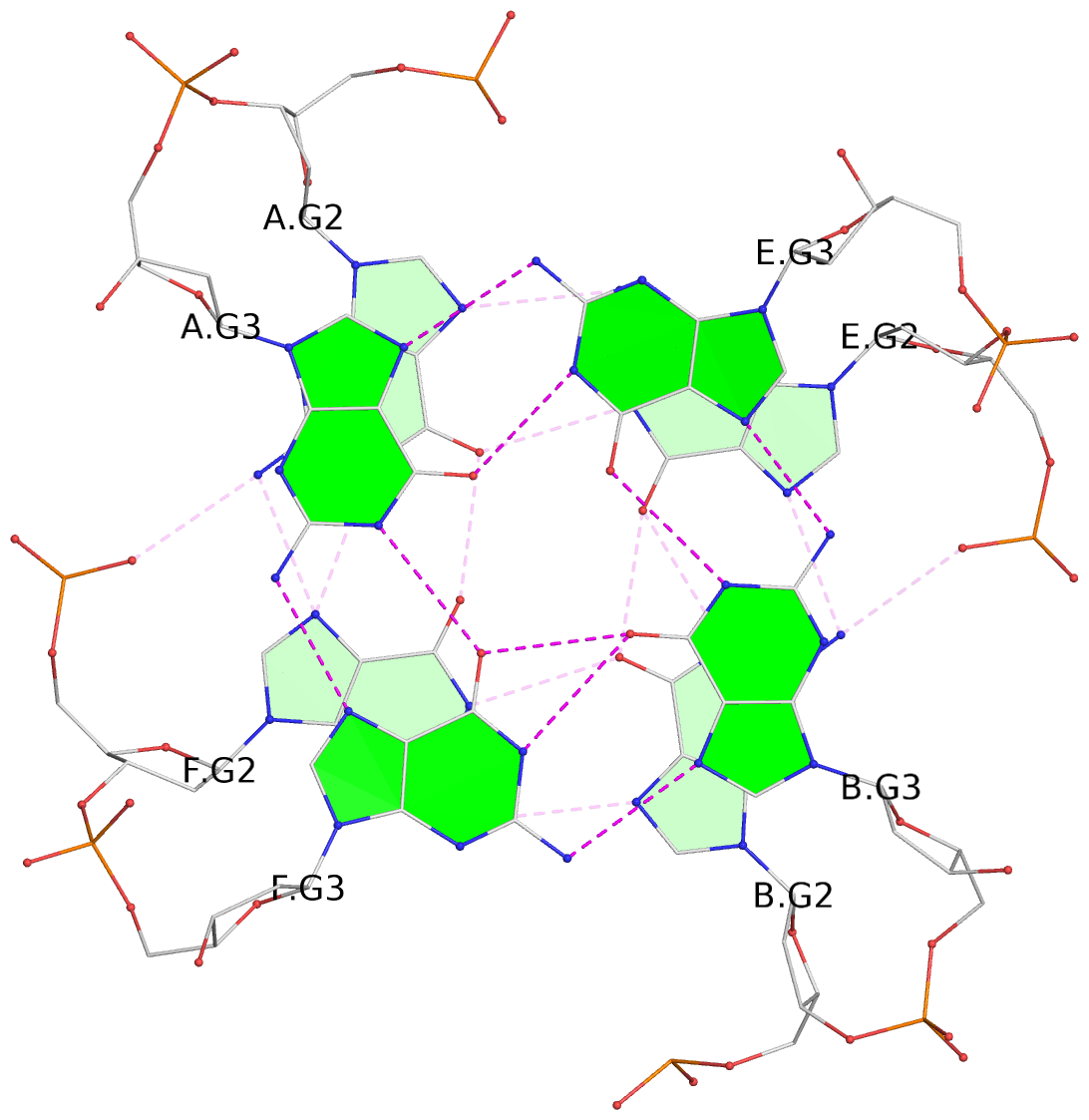
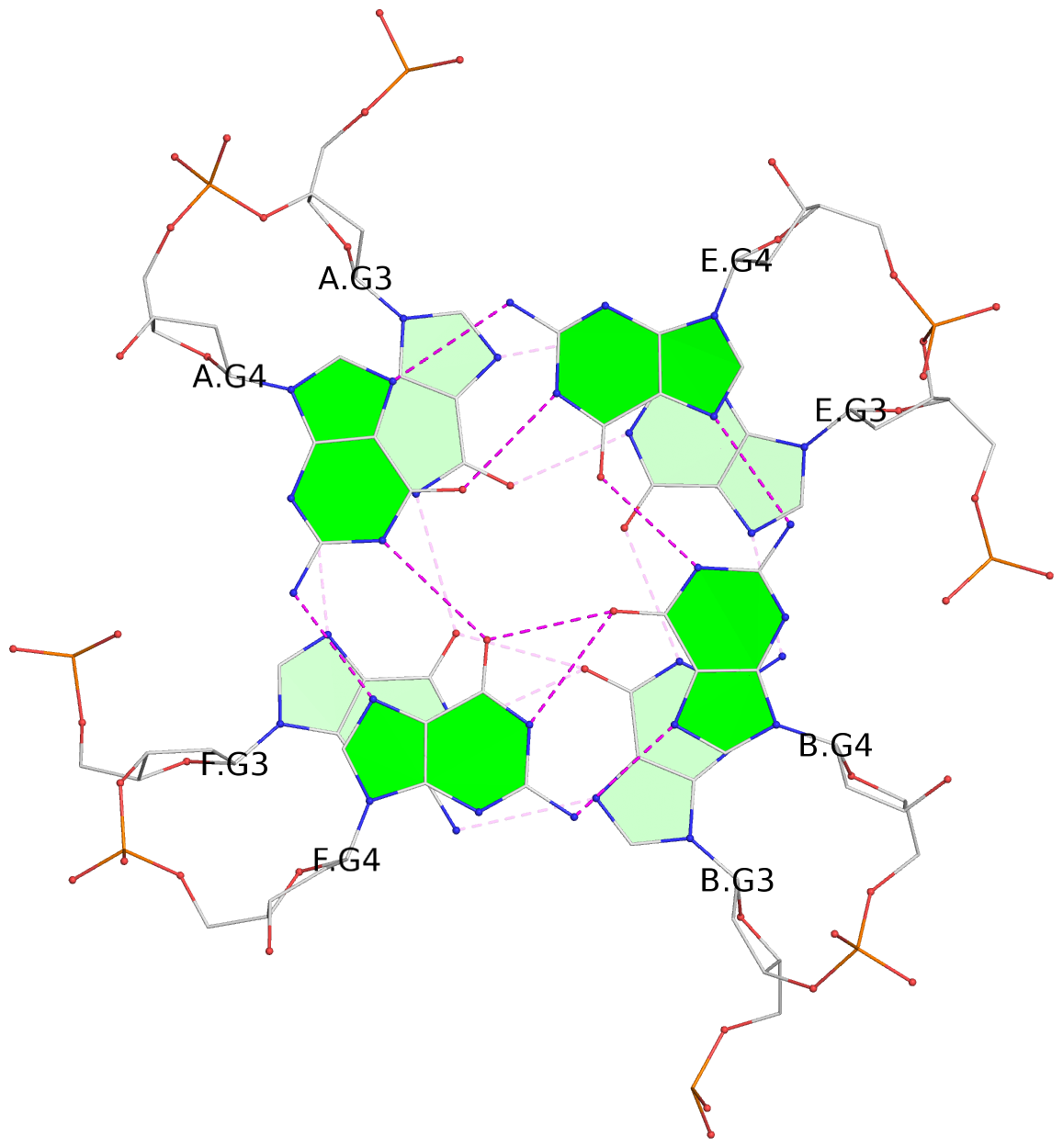
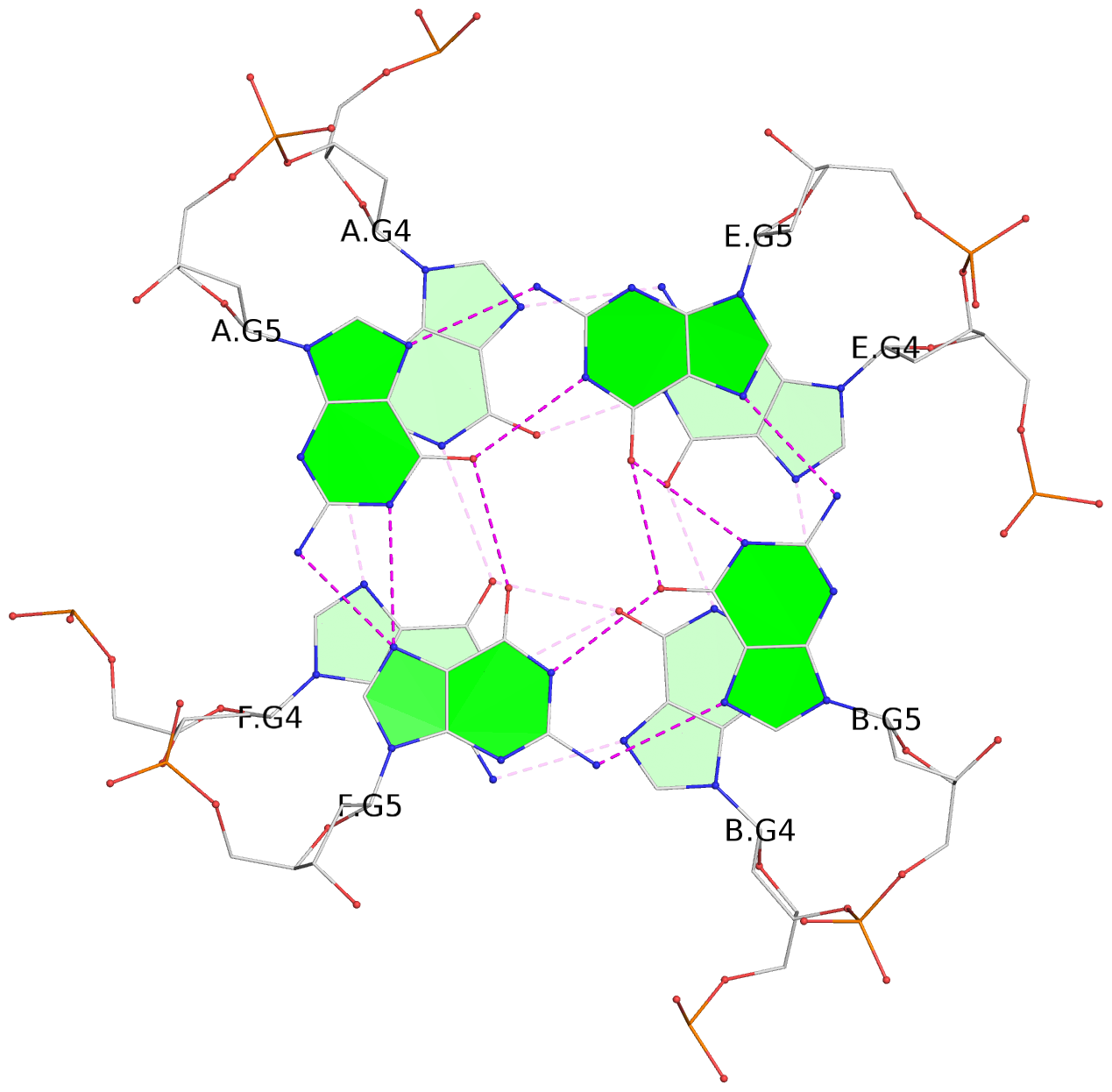
List of 4 G-tetrads
1 glyco-bond=---- sugar=-3.3 groove=---- planarity=0.223 type=other nts=4 GGGG A.DG2,F.DG2,B.DG2,E.DG2 2 glyco-bond=---- sugar=-..- groove=---- planarity=0.430 type=other nts=4 GGGG A.DG3,F.DG3,B.DG3,E.DG3 3 glyco-bond=---- sugar=-.-- groove=---- planarity=0.313 type=saddle nts=4 GGGG A.DG4,F.DG4,B.DG4,E.DG4 4 glyco-bond=---- sugar=---- groove=---- planarity=0.253 type=bowl-2 nts=4 GGGG A.DG5,F.DG5,B.DG5,E.DG5
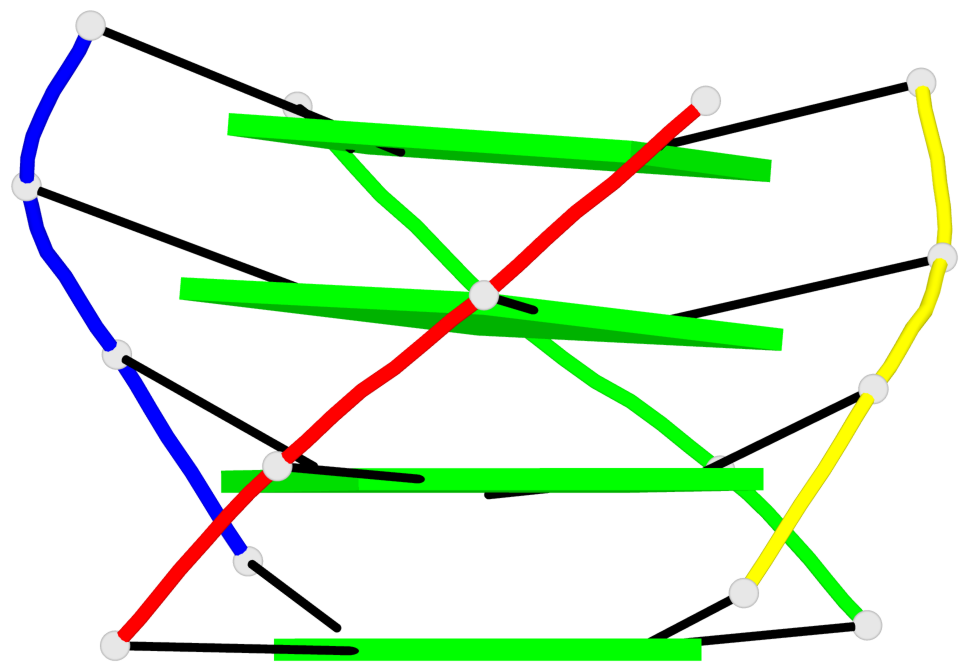
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 4 G-tetrad layers, inter-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.