Detailed DSSR results for the G-quadruplex: PDB entry 2kaz
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 2kaz
- Class
- DNA
- Method
- NMR
- Summary
- Folding topology of a bimolecular DNA quadruplex containing a stable mini-hairpin motif within the connecting loop
- Reference
- Balkwill GD, Garner TP, Williams HE, Searle MS (2009): "Folding topology of a bimolecular DNA quadruplex containing a stable mini-hairpin motif within the diagonal loop." J.Mol.Biol., 385, 1600-1615. doi: 10.1016/j.jmb.2008.11.050.
- Abstract
- We describe the NMR structural characterisation of a bimolecular anti-parallel DNA quadruplex d(G(3)ACGTAGTG(3))(2) containing an autonomously stable mini-hairpin motif inserted within the diagonal loop. A folding topology is identified that is different from that observed for the analogous d(G(3)T(4)G(3))(2) dimer with the two structures differing in the relative orientation of the diagonal loops. This appears to reflect specific base stacking interactions at the quadruplex-duplex interface that are not present in the structure with the T(4)-loop sequence. A truncated version of the bimolecular quadruplex d(G(2)ACGTAGTG(2))(2), with only two core G-tetrads, is less stable and forms a heterogeneous mixture of three 2-fold symmetric quadruplexes with different loop arrangements. We demonstrate that the nature of the loop sequence, its ability to form autonomously stable structure, the relative stabilities of the hairpin loop and core quadruplex, and the ability to form favourable stacking interactions between these two motifs are important factors in controlling DNA G-quadruplex topology.
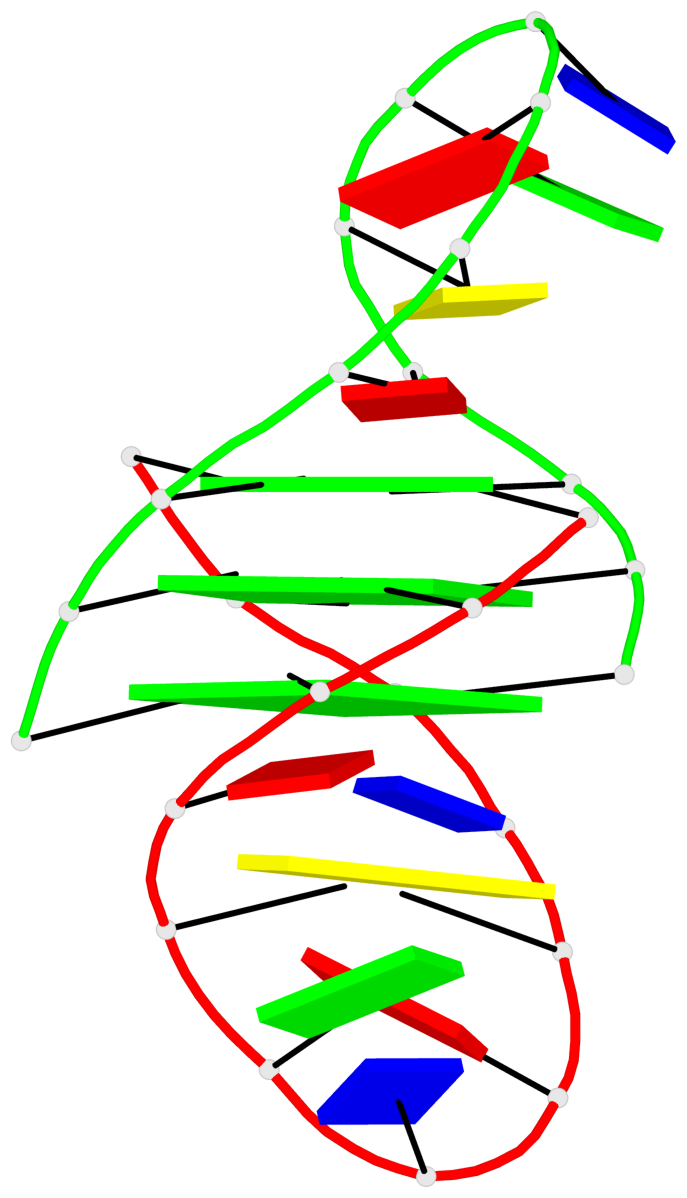
- G4 notes
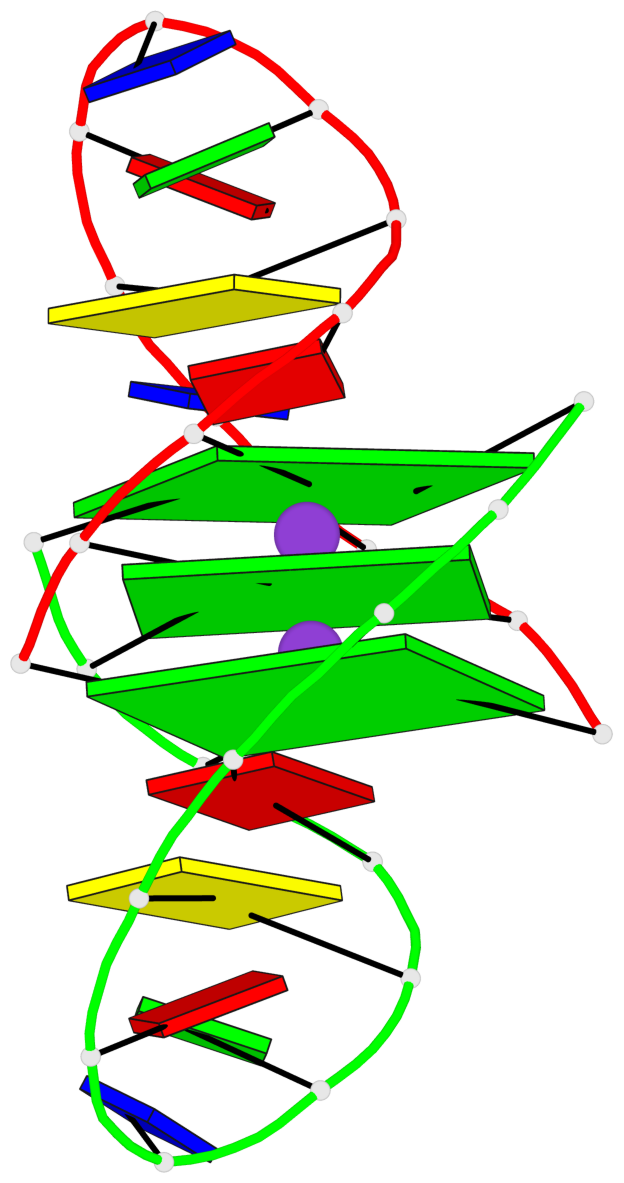
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, (2+2), UUDD
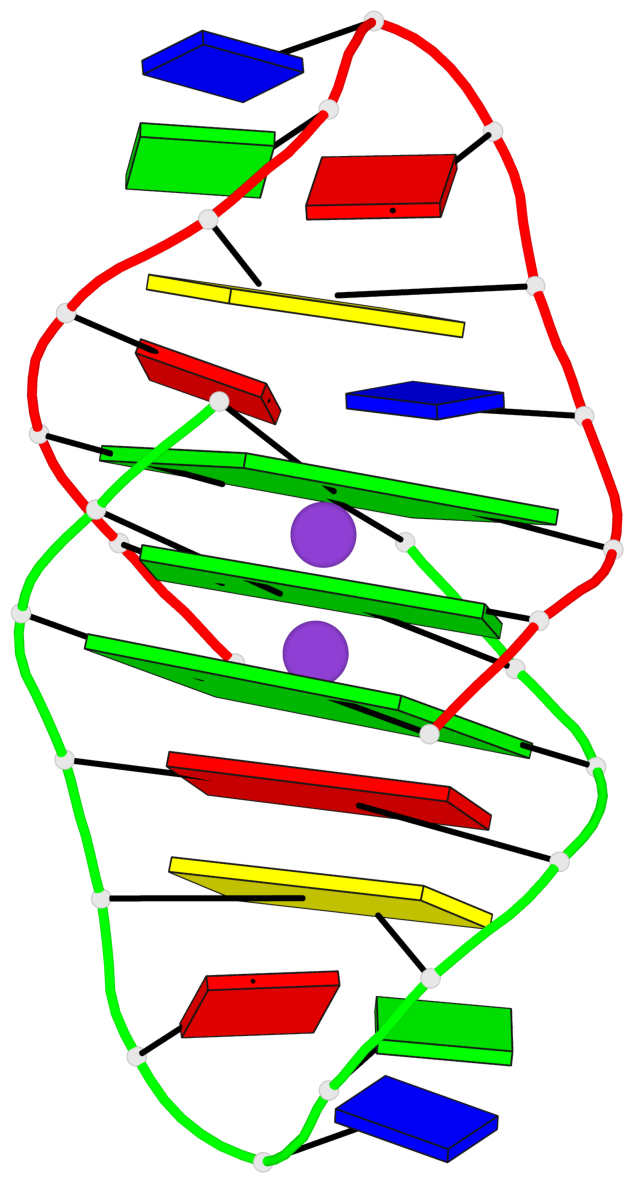
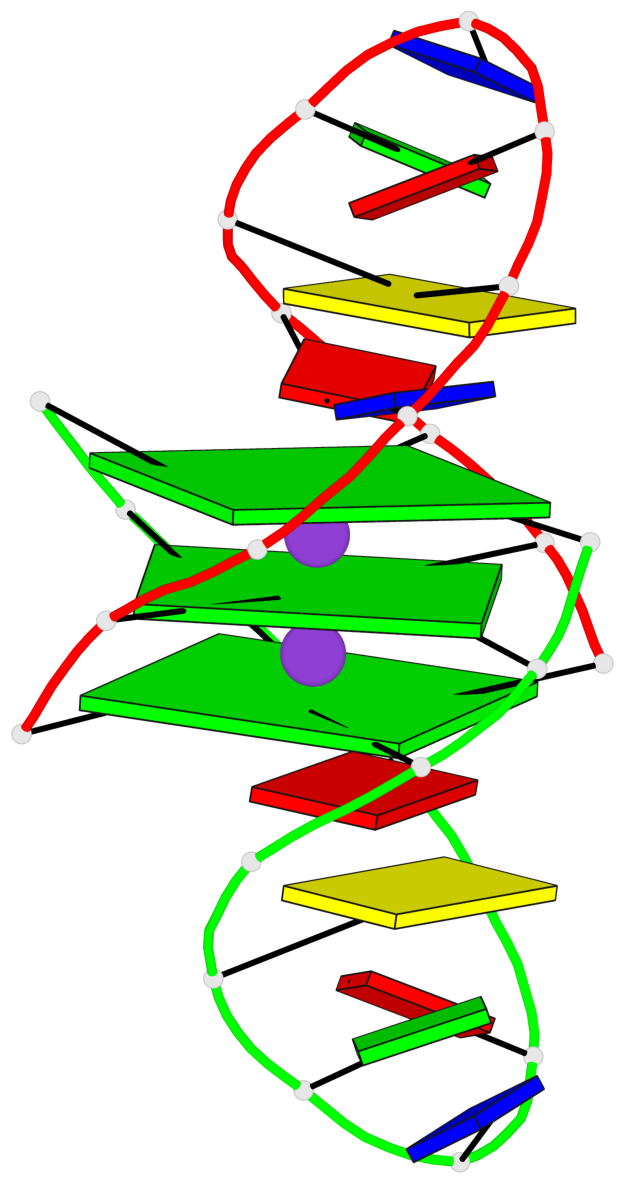
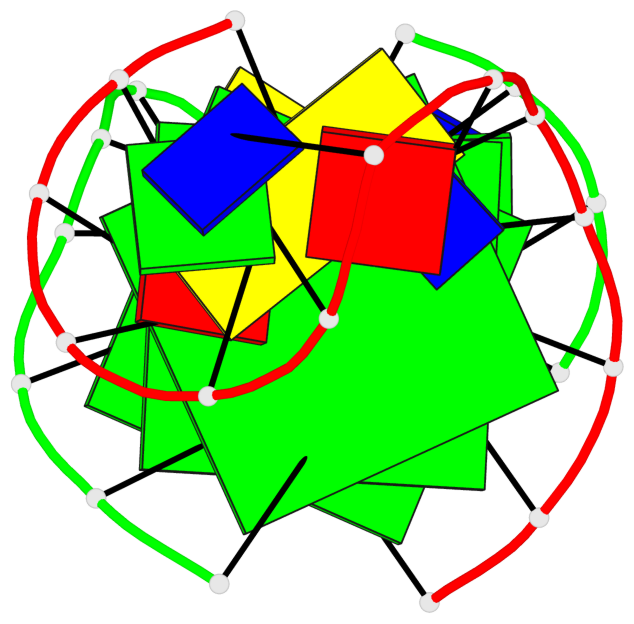
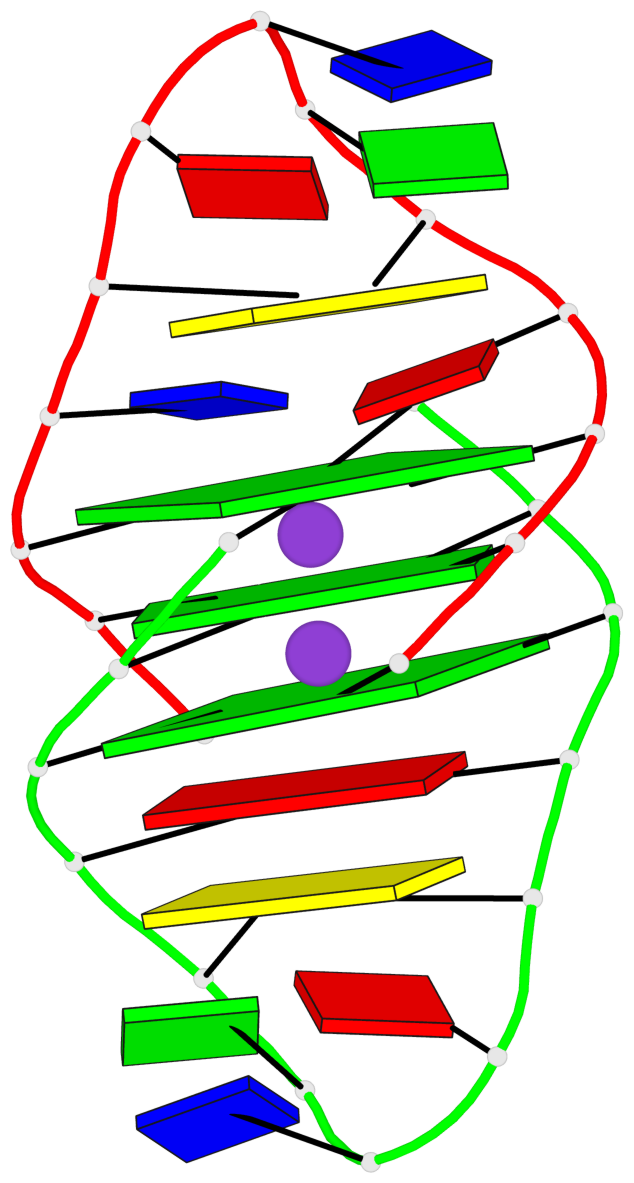
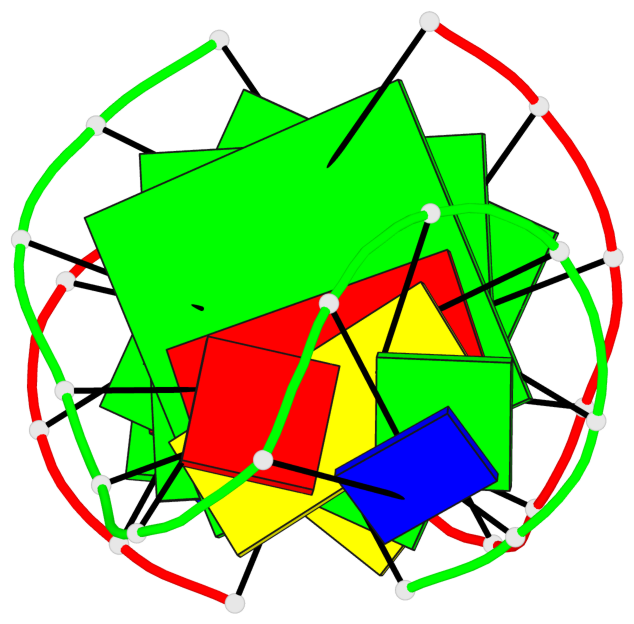
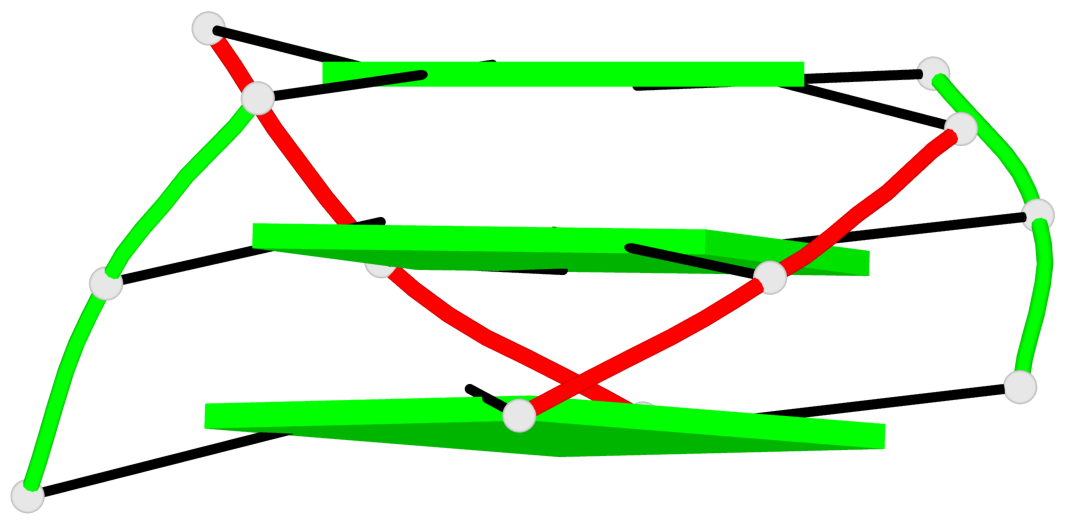
Base-block schematics in six views
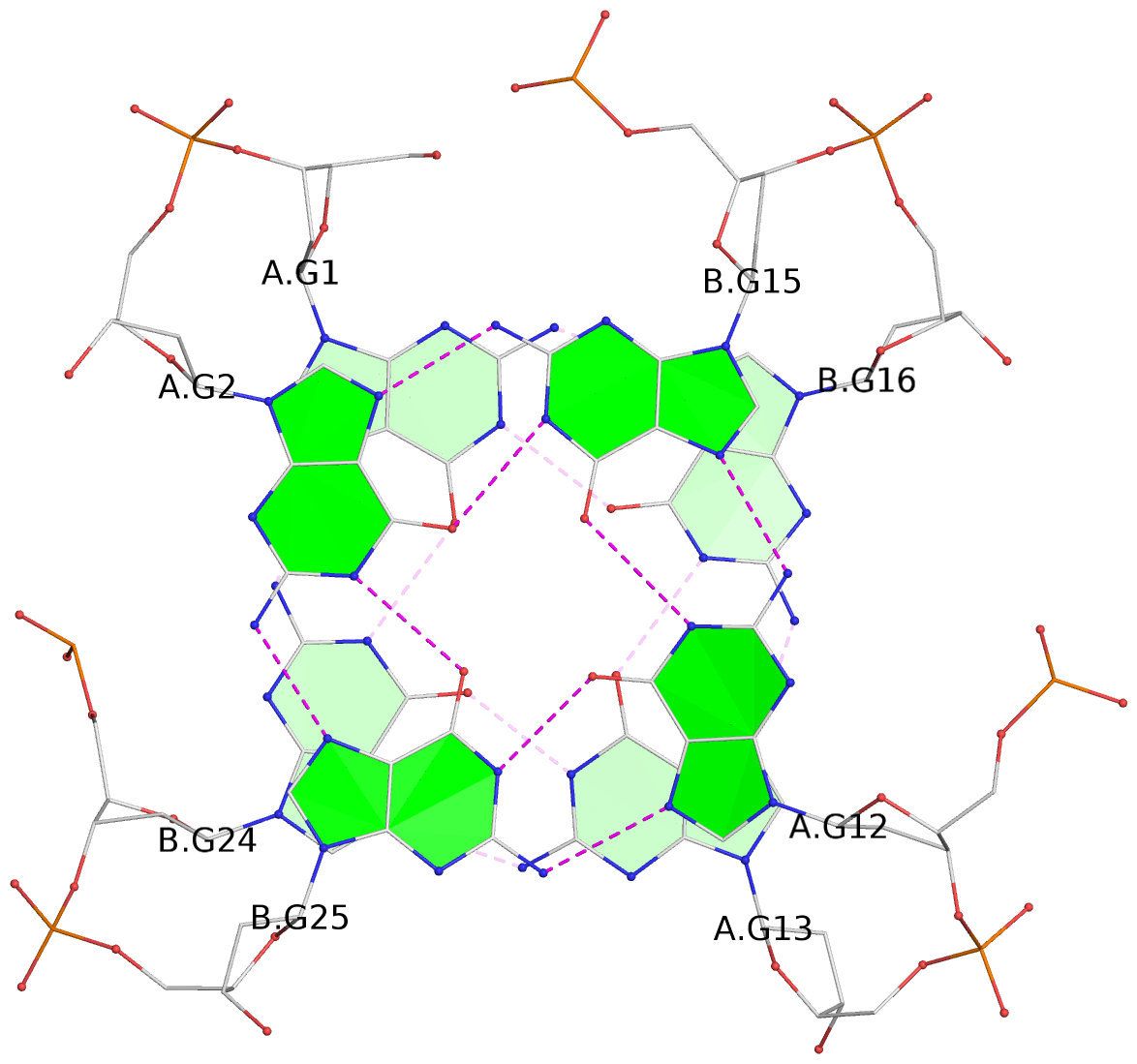
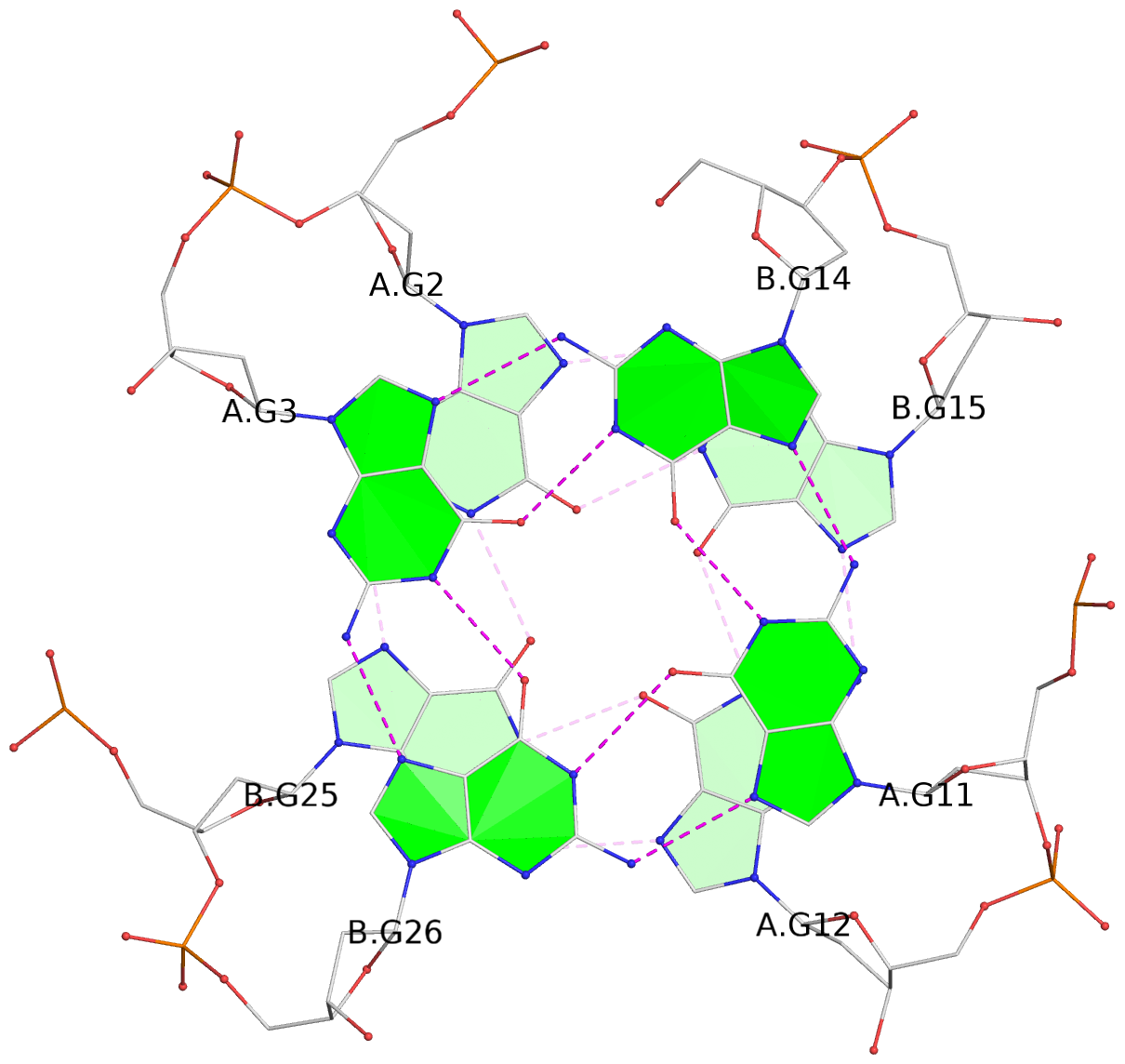
List of 3 G-tetrads
1 glyco-bond=ss-- sugar=---- groove=-w-n planarity=0.168 type=other nts=4 GGGG A.DG1,B.DG24,A.DG13,B.DG16 2 glyco-bond=--ss sugar=---- groove=-w-n planarity=0.271 type=other nts=4 GGGG A.DG2,B.DG25,A.DG12,B.DG15 3 glyco-bond=--ss sugar=---3 groove=-w-n planarity=0.279 type=bowl-2 nts=4 GGGG A.DG3,B.DG26,A.DG11,B.DG14
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, inter-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.