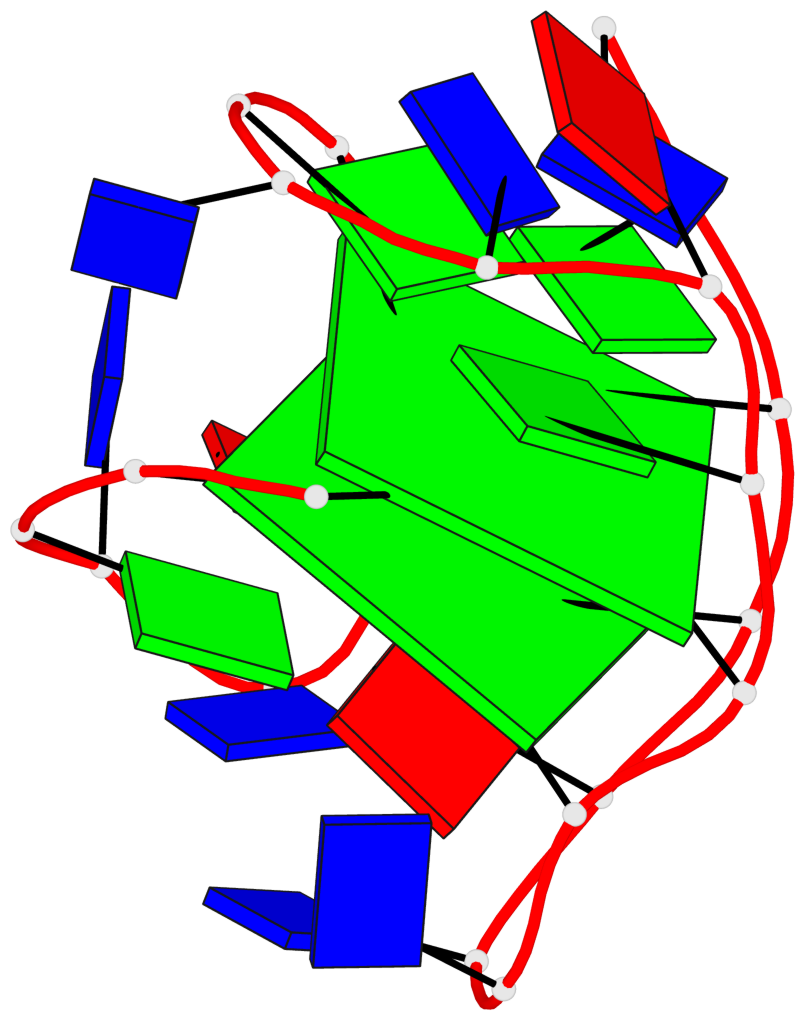
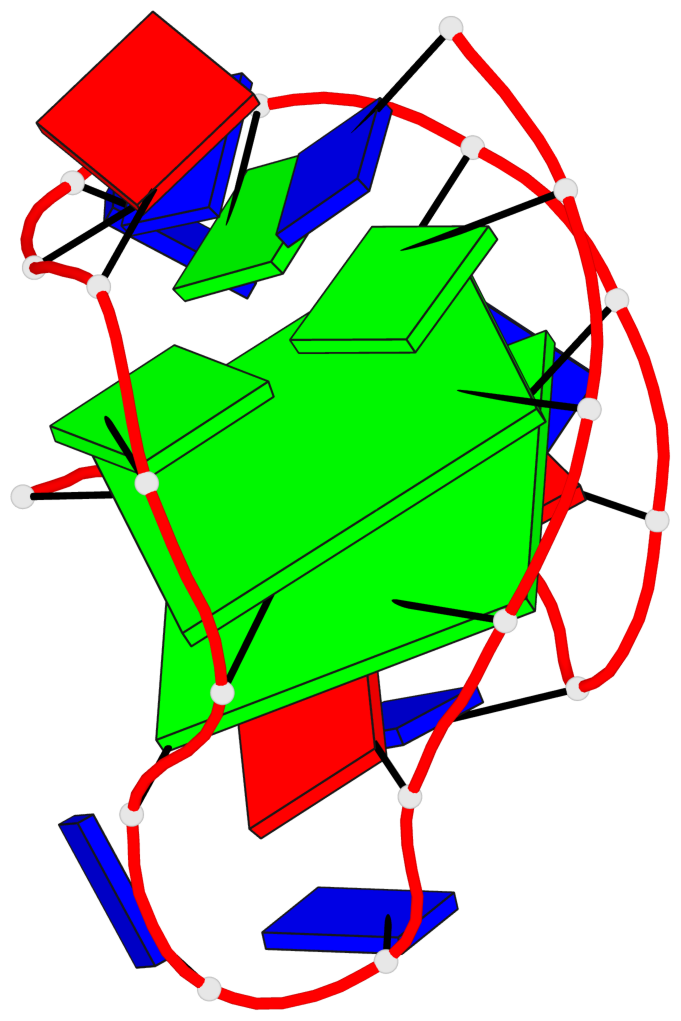
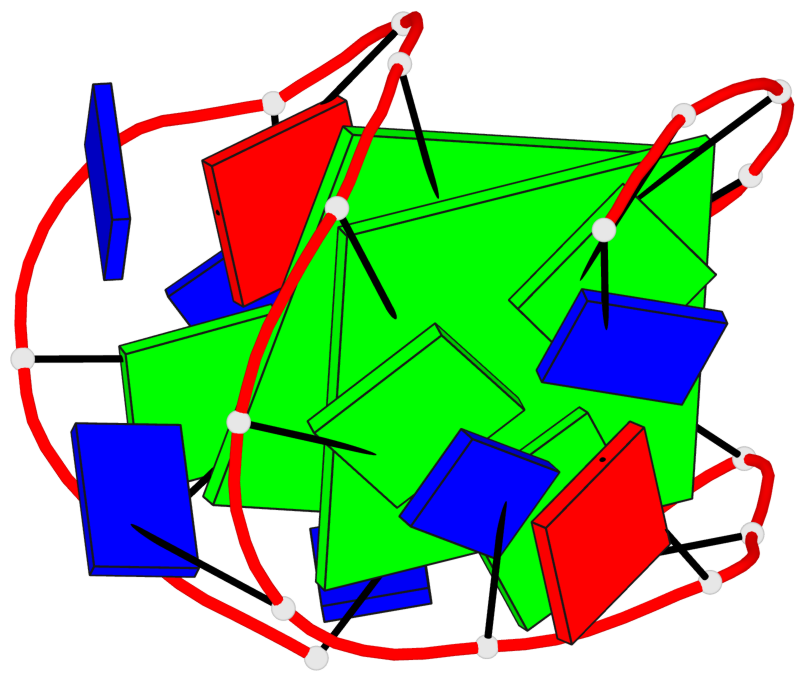
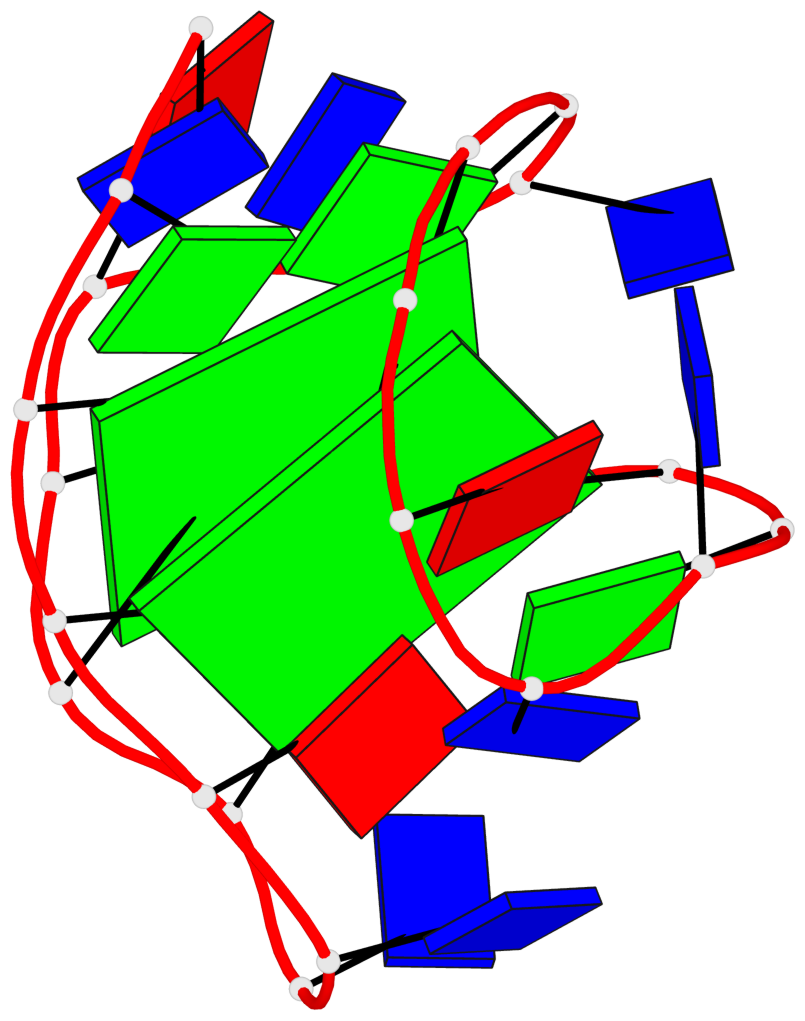
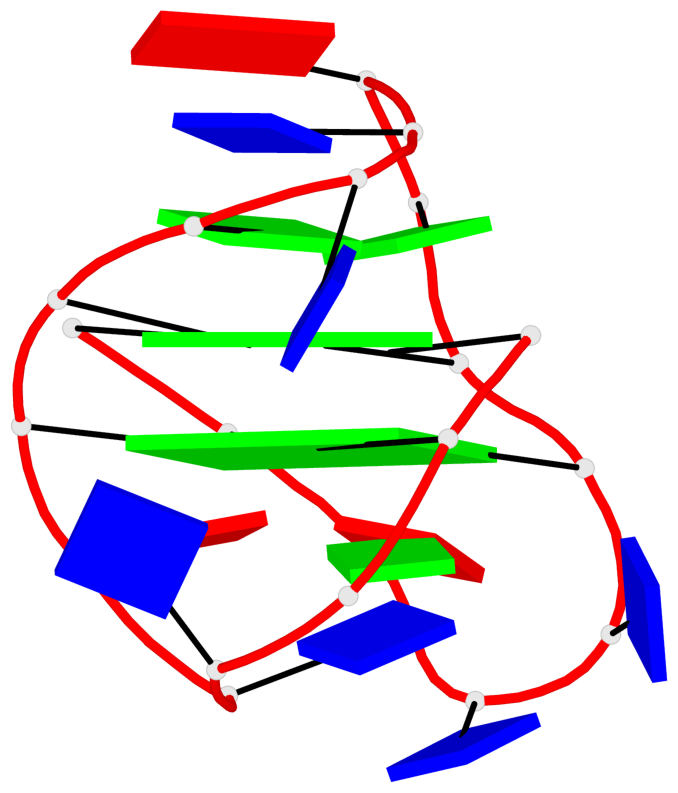
Detailed DSSR results for the G-quadruplex: PDB entry 2kf7
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 2kf7
- Class
- DNA
- Method
- NMR
- Summary
- Structure of a two-g-tetrad basket-type intramolecular g-quadruplex formed by human telomeric repeats in k+ solution (with g7-to-brg substitution)
- Reference
- Lim KW, Amrane S, Bouaziz S, Xu W, Mu Y, Patel DJ, Luu KN, Phan AT (2009): "Structure of the human telomere in K+ solution: a stable basket-type G-quadruplex with only two G-tetrad layers." J.Am.Chem.Soc., 131, 4301-4309. doi: 10.1021/ja807503g.
- Abstract
- Previously, it has been reported that human telomeric DNA sequences could adopt in different experimental conditions four different intramolecular G-quadruplexes each involving three G-tetrad layers, namely, Na(+) solution antiparallel-stranded basket form, K(+) crystal parallel-stranded propeller form, K(+) solution (3 + 1) Form 1, and K(+) solution (3 + 1) Form 2. Here we present a new intramolecular G-quadruplex adopted by a four-repeat human telomeric sequence in K(+) solution (Form 3). This structure is a basket-type G-quadruplex with only two G-tetrad layers: loops are successively edgewise, diagonal, and edgewise; glycosidic conformations of guanines are syn x syn x anti x anti around each tetrad. Each strand of the core has both a parallel and an antiparallel adjacent strands; there are one narrow, one wide, and two medium grooves. Despite the presence of only two G-tetrads in the core, this structure is more stable than the three-G-tetrad intramolecular G-quadruplexes previously observed for human telomeric sequences in K(+) solution. Detailed structural elucidation of Form 3 revealed extensive base pairing and stacking in the loops capping both ends of the G-tetrad core, which might explain the high stability of the structure. This novel structure highlights the conformational heterogeneity of human telomeric DNA. It establishes a new folding principle for G-quadruplexes and suggests new loop sequences and structures for targeting in human telomeric DNA.
- G4 notes
- 2 G-tetrads, 1 G4 helix, 1 G4 stem, 2(-LwD+Ln), basket(2+2), UDDU
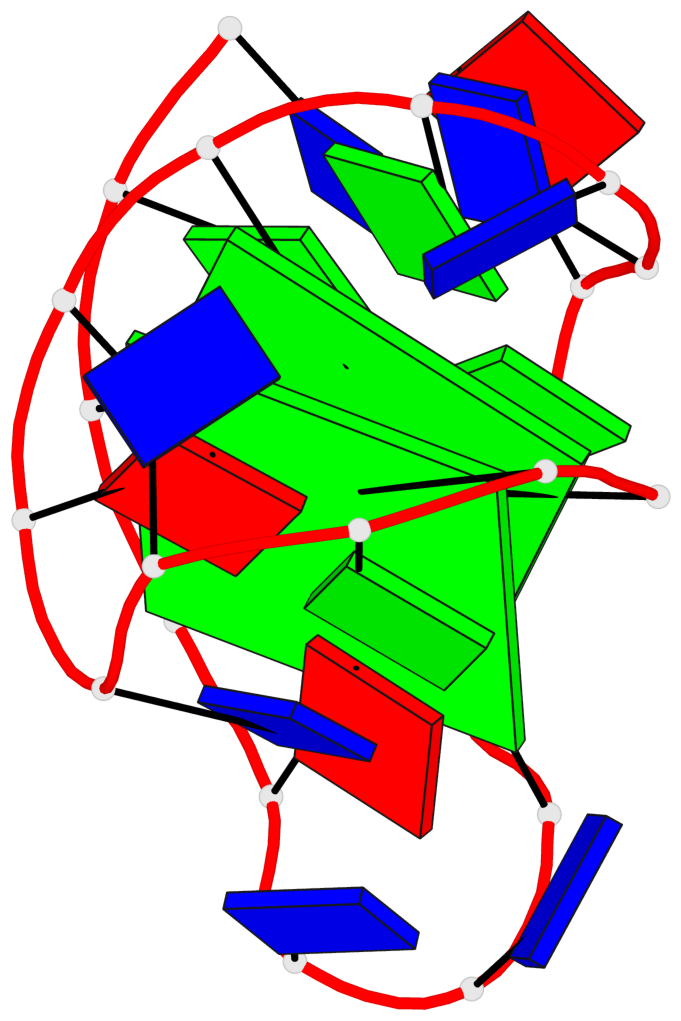
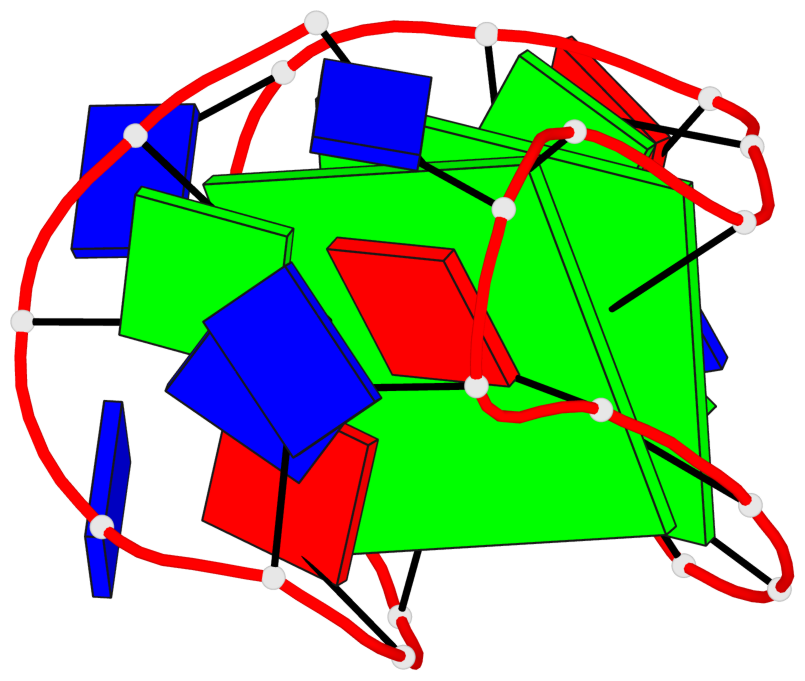
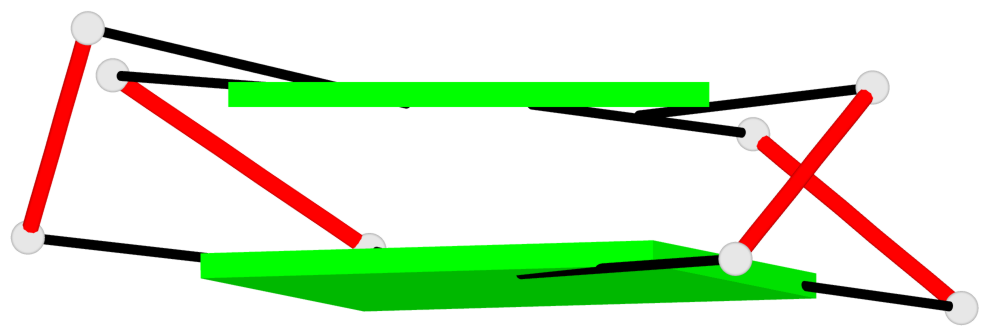
Base-block schematics in six views
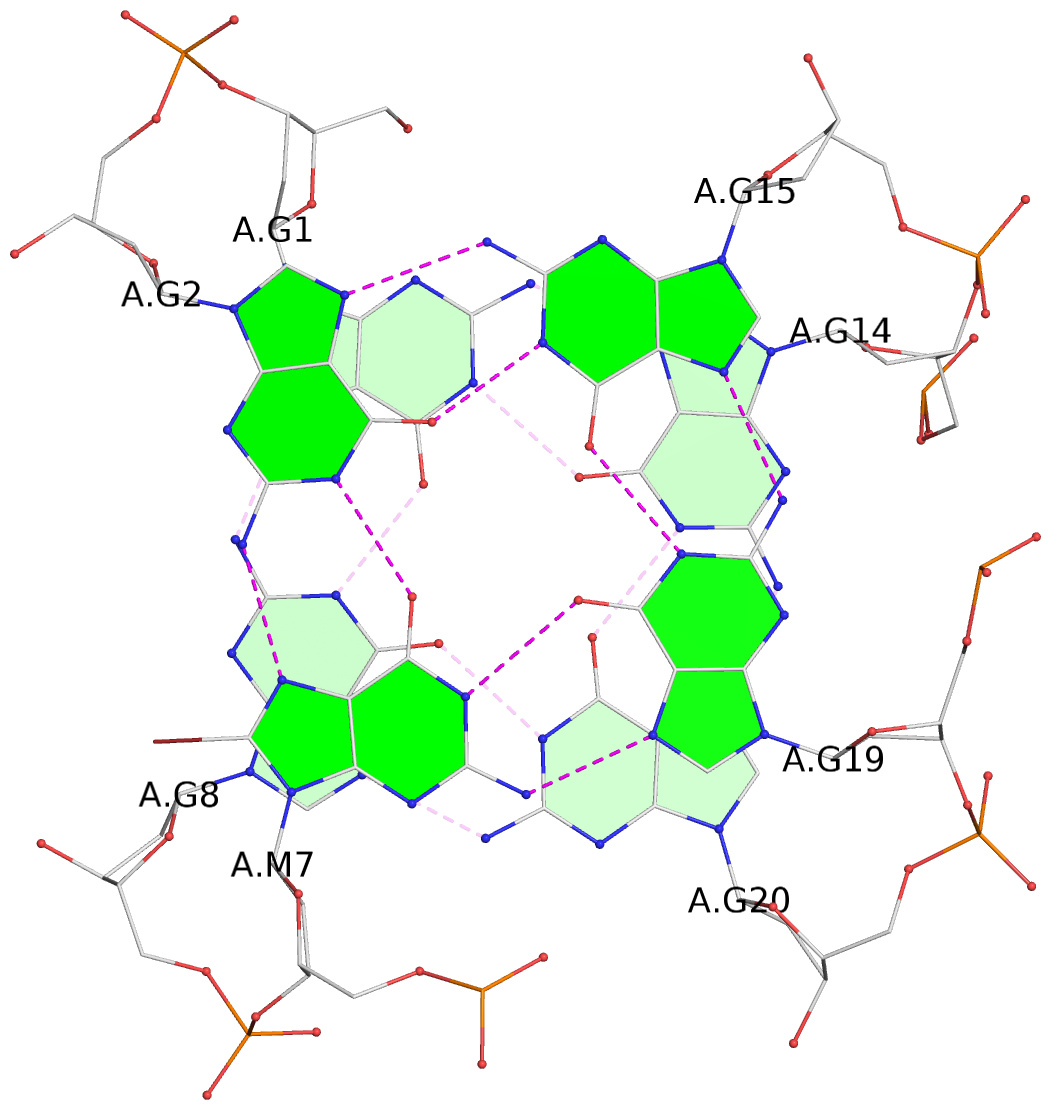
List of 2 G-tetrads
1 glyco-bond=s--s sugar=---- groove=w-n- planarity=0.259 type=other nts=4 GGGG A.DG1,A.DG8,A.DG20,A.DG14 2 glyco-bond=-ss- sugar=---- groove=w-n- planarity=0.160 type=planar nts=4 GgGG A.DG2,A.BGM7,A.DG19,A.DG15
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 2 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.