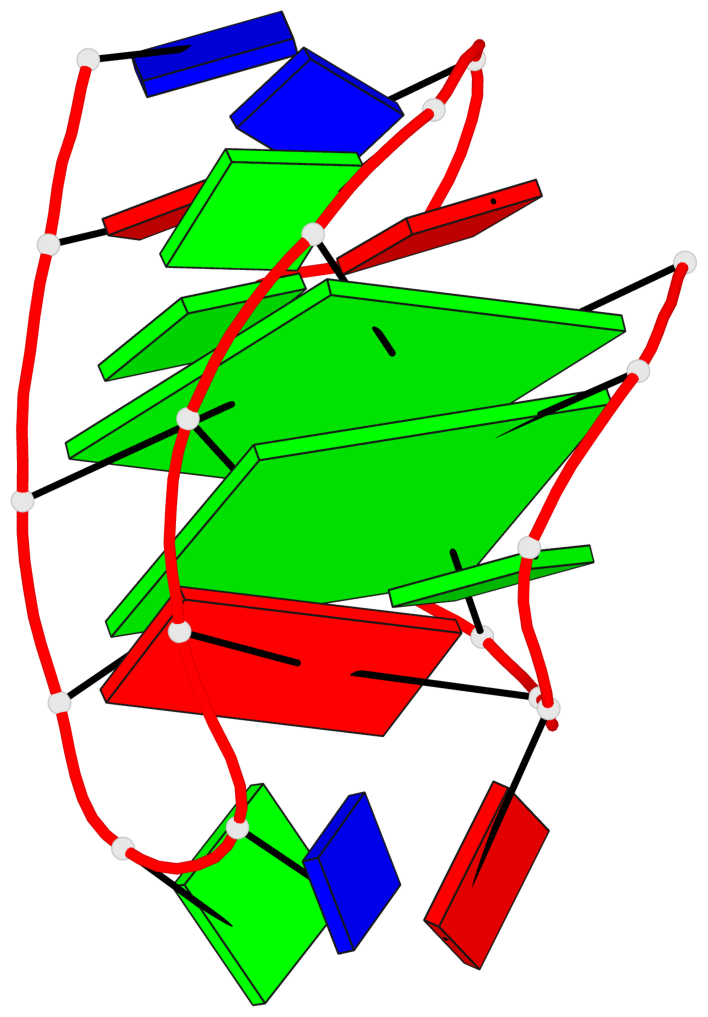
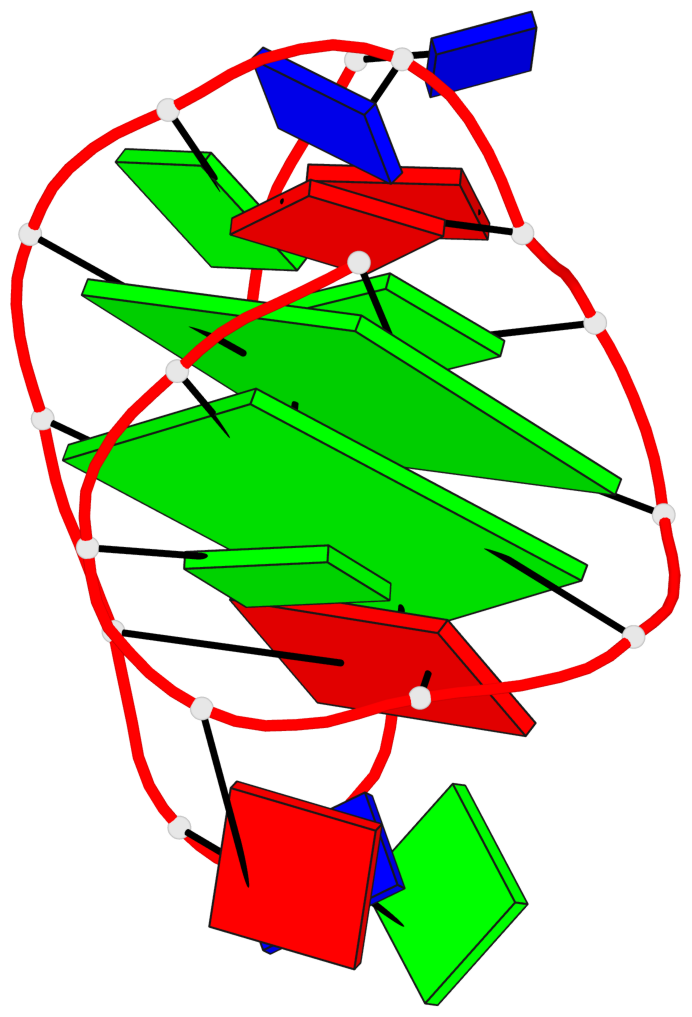
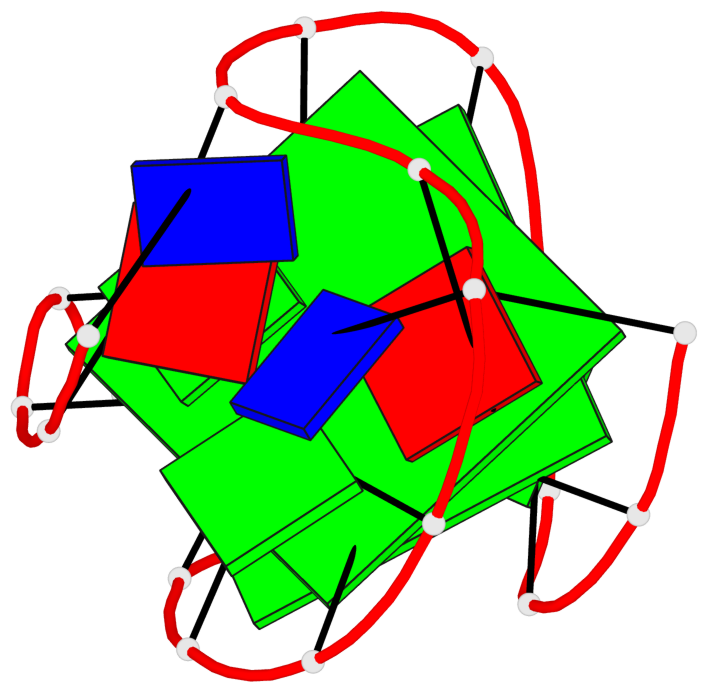
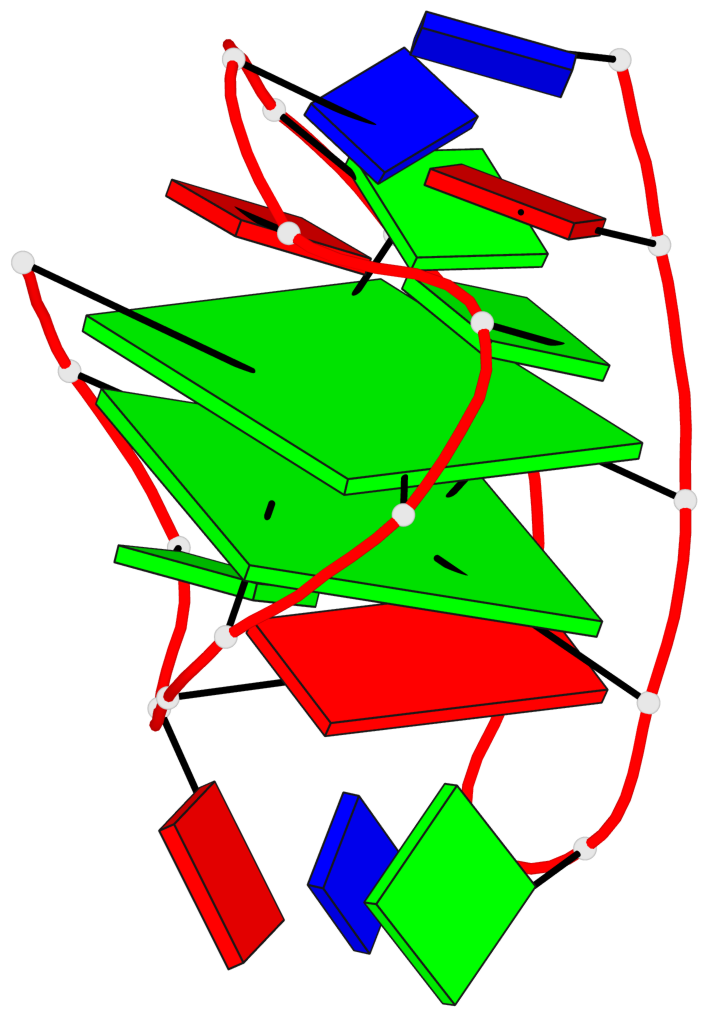
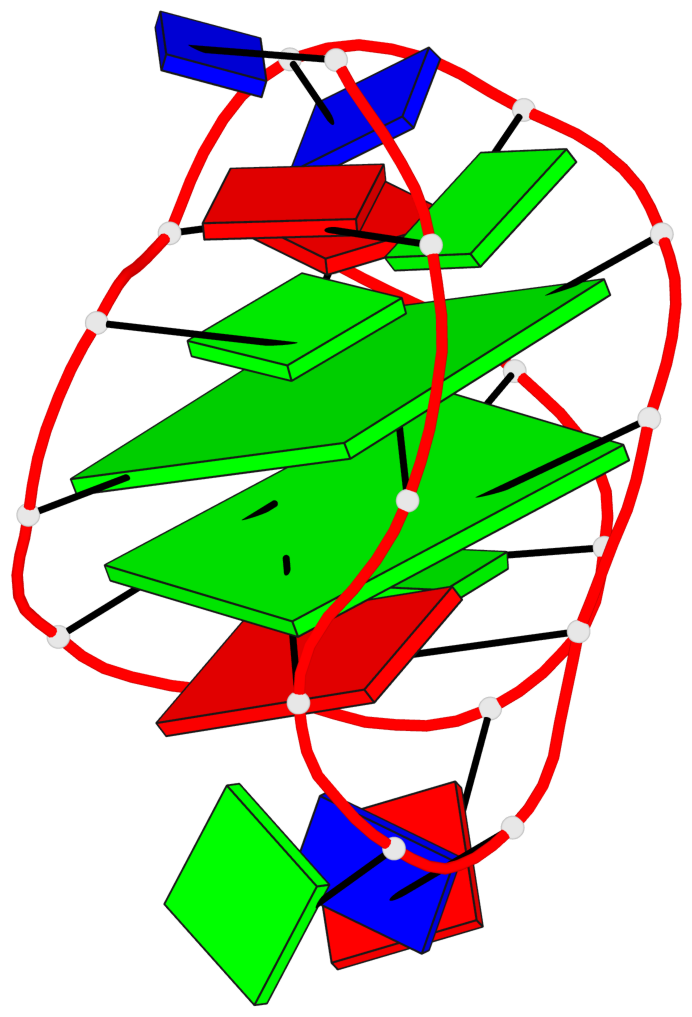
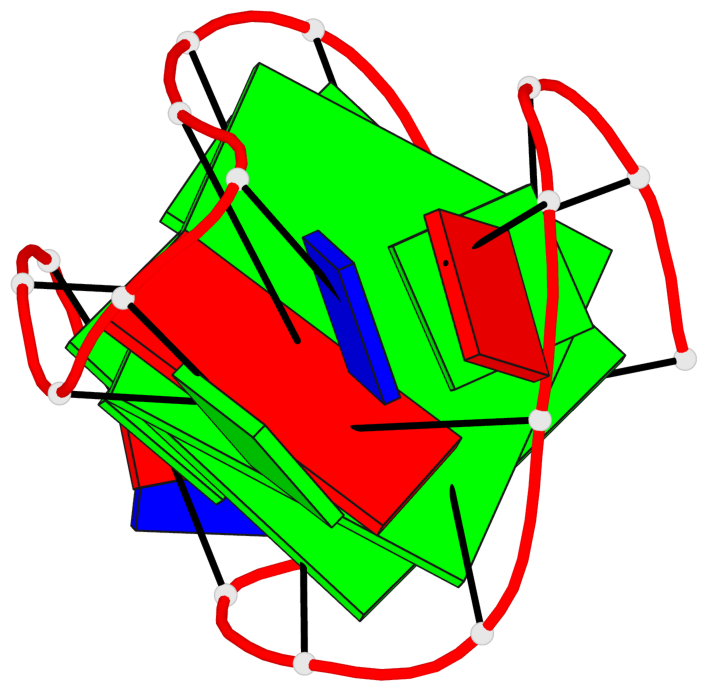
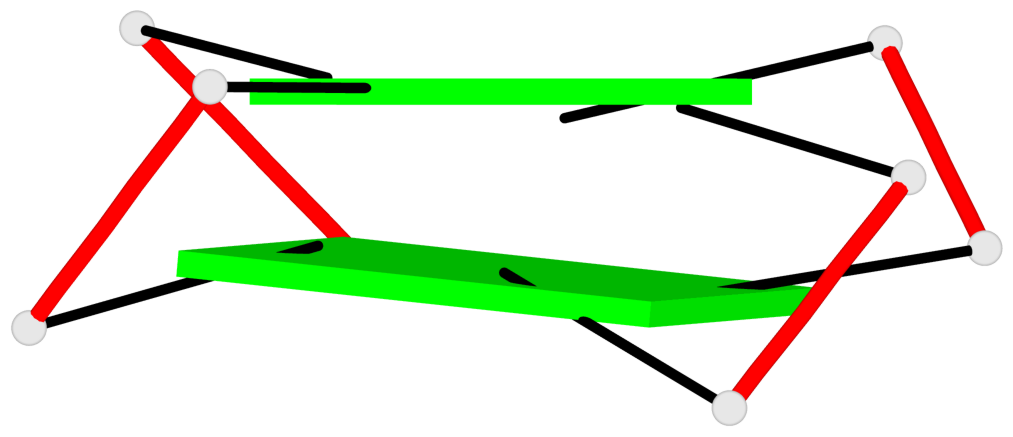
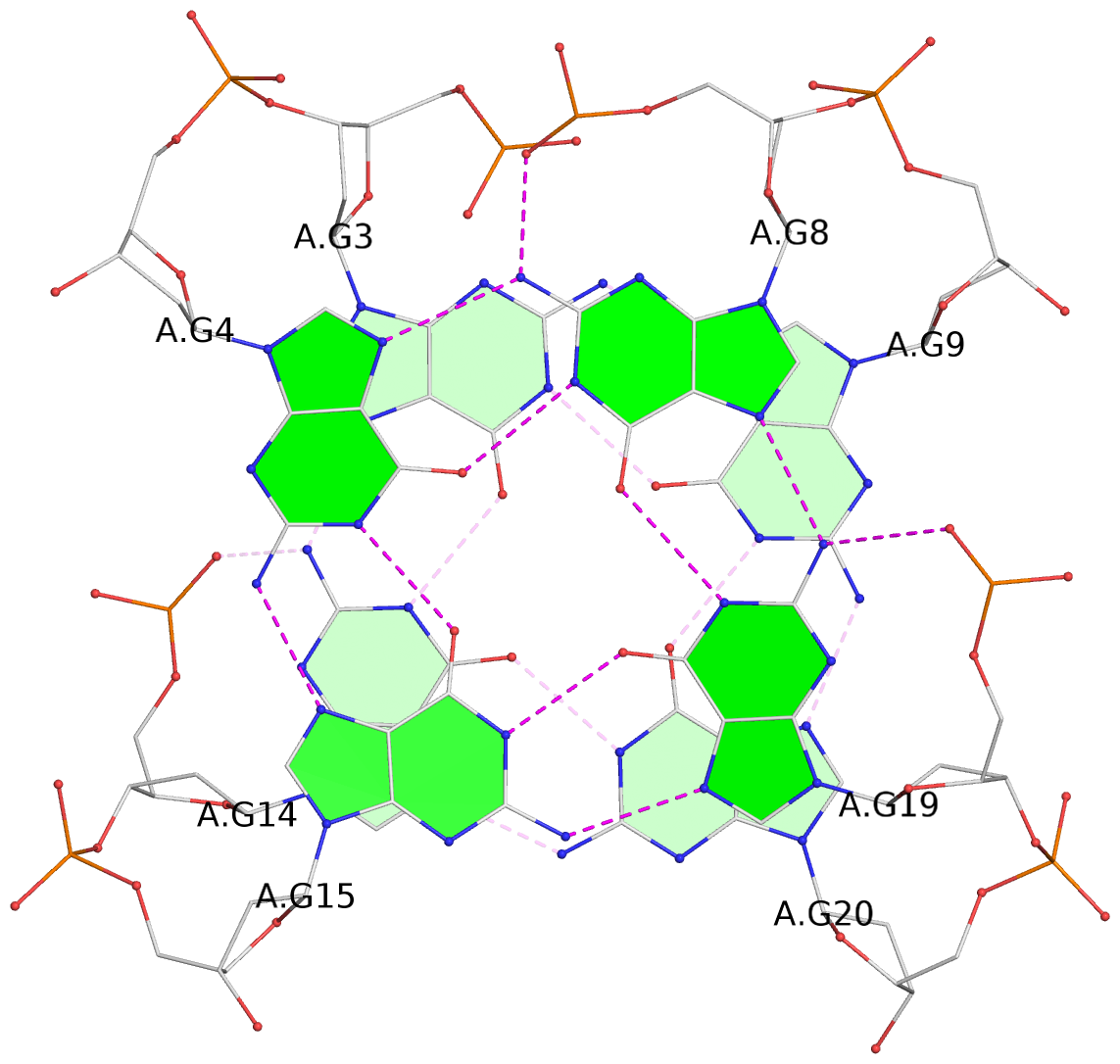
Detailed DSSR results for the G-quadruplex: PDB entry 2kow
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 2kow
- Class
- DNA
- Method
- NMR
- Summary
- Structure of a two-g-tetrad basket-type intramolecular g-quadruplex formed by giardia telomeric repeat d(taggg)4 in k+ solution (with g18-to-ino substitution)
- Reference
- Hu L, Lim KW, Bouaziz S, Phan AT (2009): "Giardia Telomeric Sequence d(TAGGG)(4) Forms Two Intramolecular G-Quadruplexes in K(+) Solution: Effect of Loop Length and Sequence on the Folding Topology." J.Am.Chem.Soc., 131, 16824-16831. doi: 10.1021/ja905611c.
- Abstract
- Recently, it has been shown that in K(+) solution the human telomeric sequence d[TAGGG(TTAGGG)(3)] forms a (3 + 1) intramolecular G-quadruplex, while the Bombyx mori telomeric sequence d[TAGG(TTAGG)(3)], which differs from the human counterpart only by one G deletion in each repeat, forms a chair-type intramolecular G-quadruplex, indicating an effect of G-tract length on the folding topology of G-quadruplexes. To explore the effect of loop length and sequence on the folding topology of G-quadruplexes, here we examine the structure of the four-repeat Giardia telomeric sequence d[TAGGG(TAGGG)(3)], which differs from the human counterpart only by one T deletion within the non-G linker in each repeat. We show by NMR that this sequence forms two different intramolecular G-quadruplexes in K(+) solution. The first one is a novel basket-type antiparallel-stranded G-quadruplex containing two G-tetrads, a G x (A-G) triad, and two A x T base pairs; the three loops are consecutively edgewise-diagonal-edgewise. The second one is a propeller-type parallel-stranded G-quadruplex involving three G-tetrads; the three loops are all double-chain-reversal. Recurrence of several structural elements in the observed structures suggests a "cut and paste" principle for the design and prediction of G-quadruplex topologies, for which different elements could be extracted from one G-quadruplex and inserted into another.
- G4 notes
- 2 G-tetrads, 1 G4 helix, 1 G4 stem, 2(+LnD-Lw), basket(2+2), UUDD
Base-block schematics in six views
List of 2 G-tetrads
1 glyco-bond=ss-- sugar=---- groove=-w-n planarity=0.256 type=other nts=4 GGGG A.DG3,A.DG14,A.DG20,A.DG9 2 glyco-bond=--ss sugar=---- groove=-w-n planarity=0.294 type=other nts=4 GGGG A.DG4,A.DG15,A.DG19,A.DG8
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 2 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.