Detailed DSSR results for the G-quadruplex: PDB entry 2kpr
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 2kpr
- Class
- DNA
- Method
- NMR
- Summary
- Monomeric intronic human chl1 gene quadruplex DNA NMR, 17 structures
- Reference
- Kuryavyi V, Patel DJ (2010): "Solution Structure of a Unique G-Quadruplex Scaffold Adopted by a Guanosine-Rich Human Intronic Sequence." Structure, 18, 73-82. doi: 10.1016/j.str.2009.10.015.
- Abstract
- We report on the solution structure of an unprecedented intramolecular G-quadruplex formed by the guanosine-rich human chl1 intronic d(G(3)-N-G(4)-N(2)-G(4)-N-G(3)-N) 19-mer sequence in K(+)-containing solution. This G-quadruplex, composed of three stacked G-tetrads containing four syn guanines, represents a new folding topology with two unique conformational features. The first guanosine is positioned within the central G-tetrad, in contrast to all previous structures of unimolecular G-quadruplexes, where the first guanosine is part of an outermost G-tetrad. In addition, a V-shaped loop, spanning three G-tetrad planes, contains no bridging nucleotides. The G-quadruplex scaffold is stabilized by a T*G*A triple stacked over the G-tetrad at one end and an unpaired guanosine stacked over the G-tetrad at the other end. Finally, the chl1 intronic DNA G-quadruplex scaffold contains a guanosine base intercalated between an extended G-G step, a feature observed in common with the catalytic site of group I introns. This unique structural scaffold provides a highly specific platform for the future design of ligands specifically targeted to intronic G-quadruplex platforms.
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 2(-LwX+P), UD3(1+3), UDDD
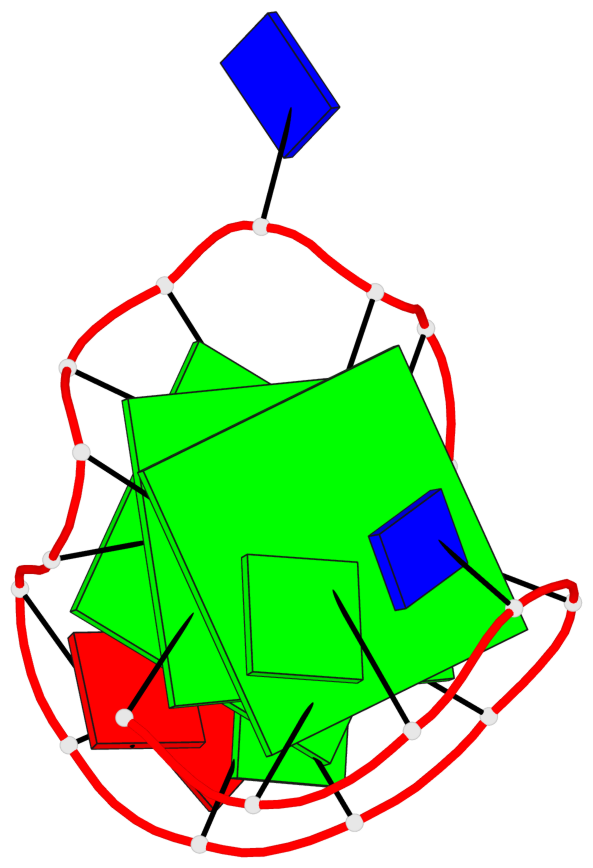
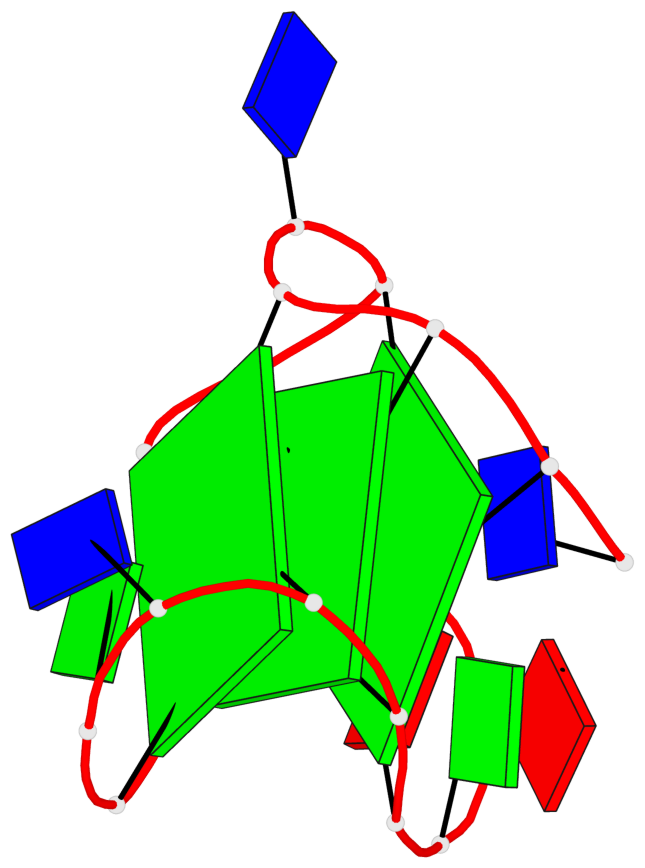
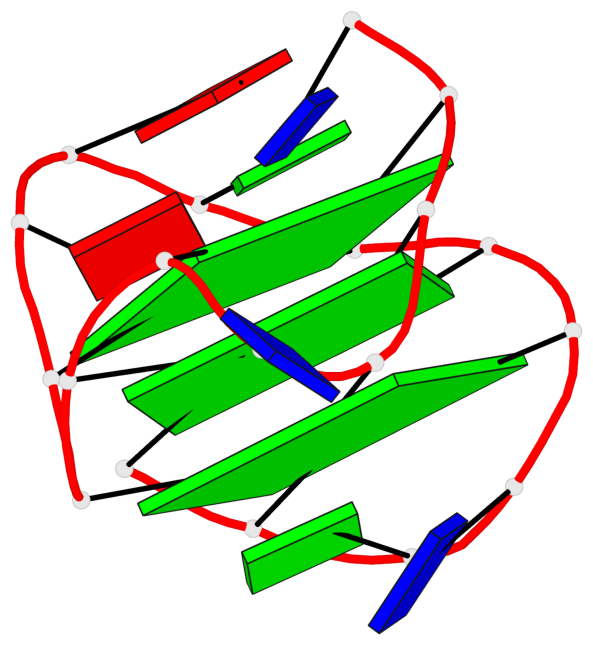
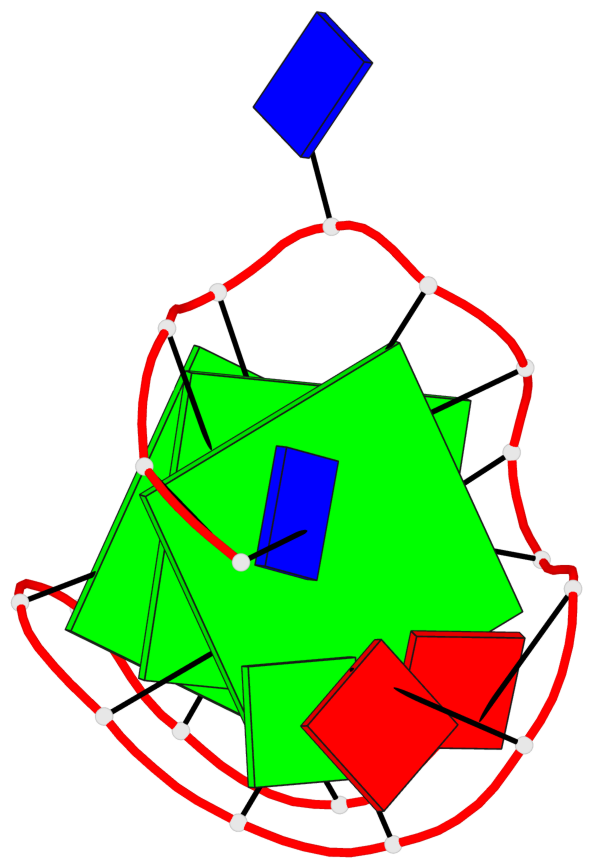
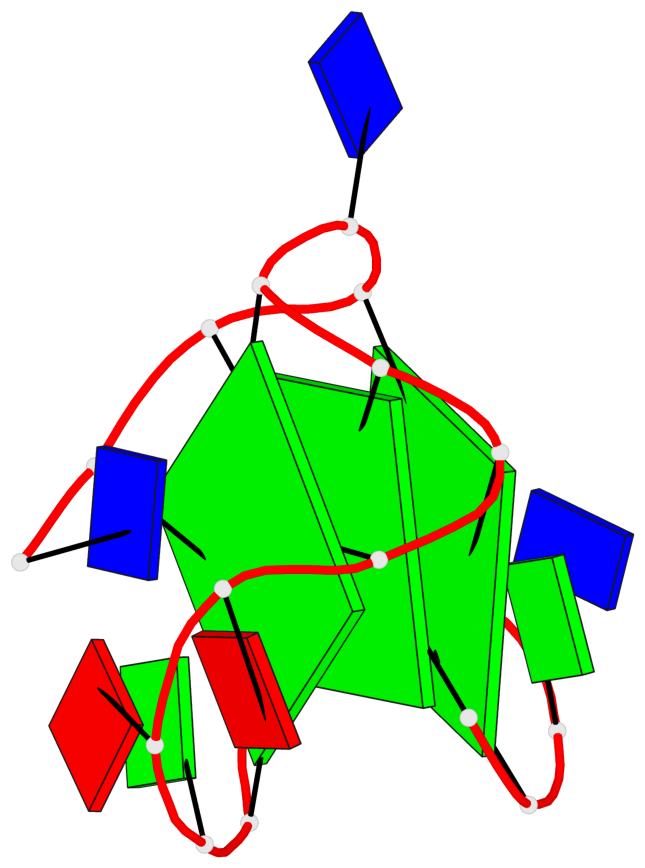
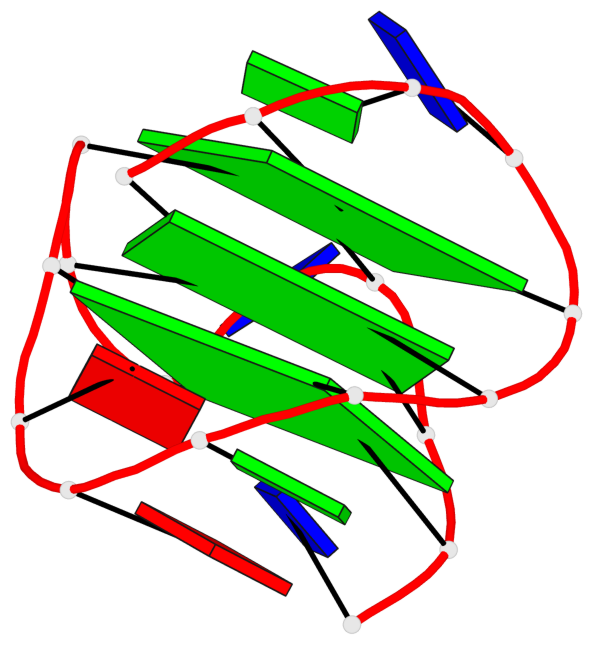
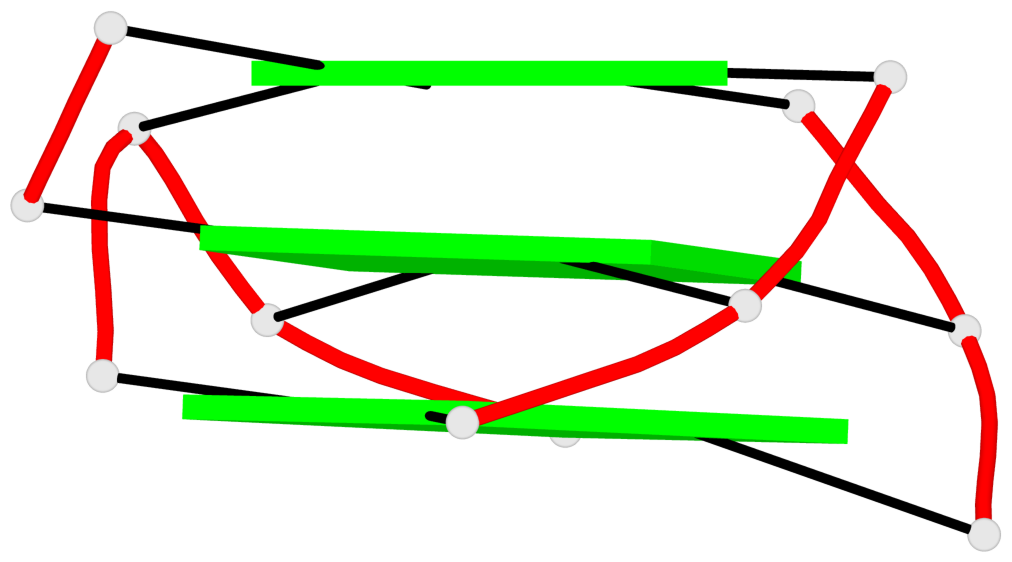
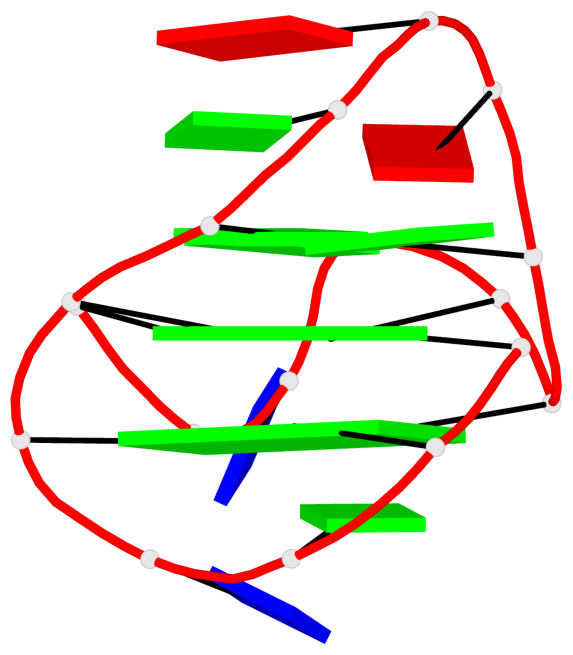
Base-block schematics in six views
List of 3 G-tetrads
1 glyco-bond=s--- sugar=---- groove=w--n planarity=0.134 type=planar nts=4 GGGG A.DG1,A.DG6,A.DG17,A.DG13 2 glyco-bond=-sss sugar=---. groove=w--n planarity=0.162 type=other nts=4 GGGG A.DG2,A.DG5,A.DG16,A.DG12 3 glyco-bond=-s-- sugar=-3-3 groove=wn-- planarity=0.133 type=planar nts=4 GGGG A.DG7,A.DG11,A.DG14,A.DG18
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 2 G-tetrad layers, 3 loops, INTRA-molecular, UDDD, hybrid-(mixed), 2(-LwX+P), UD3(1+3)
List of 2 non-stem G4-loops (including the two closing Gs)
1 type=lateral helix=#1 nts=5 GGAAG A.DG7,A.DG8,A.DA9,A.DA10,A.DG11 2 type=V-shaped helix=#1 nts=4 GGGG A.DG11,A.DG12,A.DG13,A.DG14