Detailed DSSR results for the G-quadruplex: PDB entry 2kqg
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 2kqg
- Class
- DNA
- Method
- NMR
- Summary
- A g-rich sequence within the c-kit oncogene promoter forms a parallel g-quadruplex having asymmetric g-tetrad dynamics
- Reference
- Hsu S-TD, Varnai P, Bugaut A, Reszka AP, Neidle S, Balasubramanian S (2009): "A G-rich sequence within the c-kit oncogene promoter forms a parallel G-quadruplex having asymmetric G-tetrad dynamics." J.Am.Chem.Soc., 131, 13399-13409. doi: 10.1021/ja904007p.
- Abstract
- Guanine-rich DNA sequences with the ability to form quadruplex structures are enriched in the promoter regions of protein-coding genes, particularly those of proto-oncogenes. G-quadruplexes are structurally polymorphic and their folding topologies can depend on the sample conditions. We report here on a structural study using solution state NMR spectroscopy of a second G-quadruplex-forming motif (c-kit2) that has been recently identified in the promoter region of the c-kit oncogene. In the presence of potassium ions, c-kit2 exists as an ensemble of structures that share the same parallel-stranded propeller-type conformations. Subtle differences in structural dynamics have been identified using hydrogen-deuterium exchange experiments by NMR spectroscopy, suggesting the coexistence of at least two structurally similar but dynamically distinct substates, which undergo slow interconversion on the NMR timescale.
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(-P-P-P), parallel(4+0), UUUU
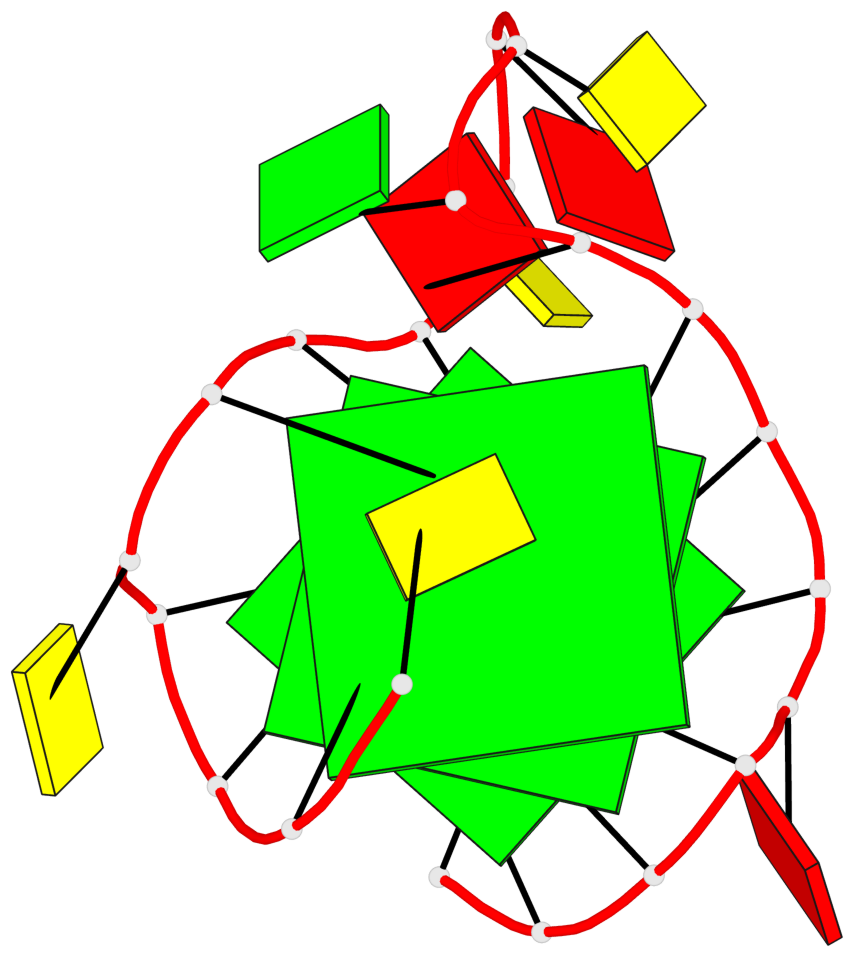
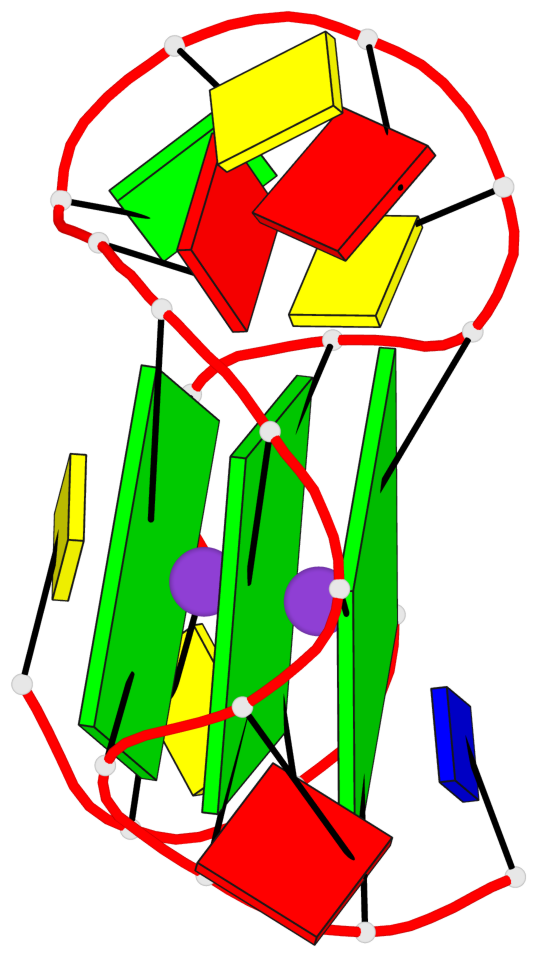
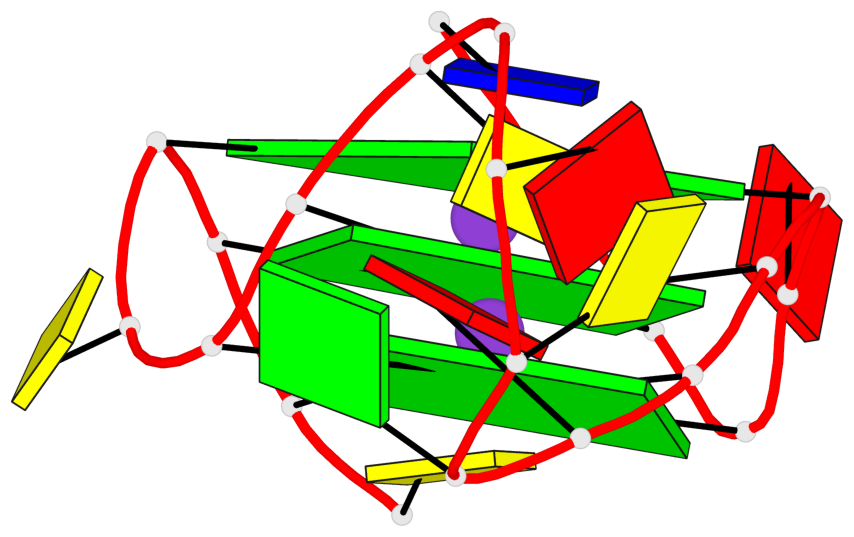
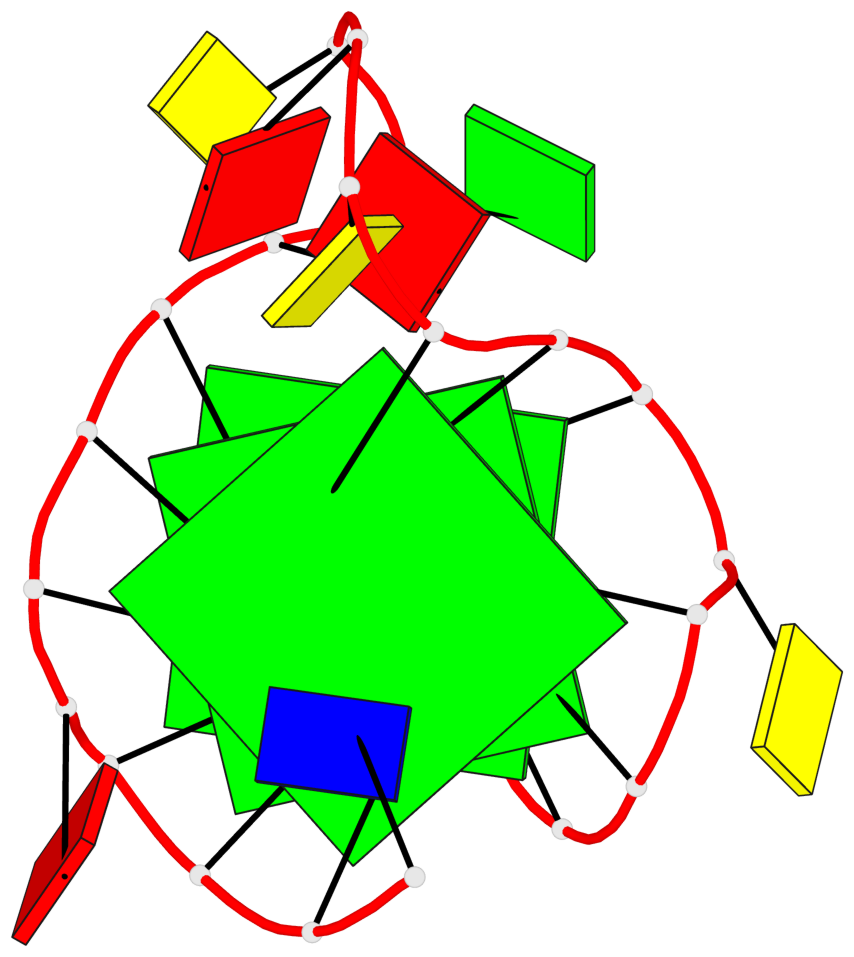
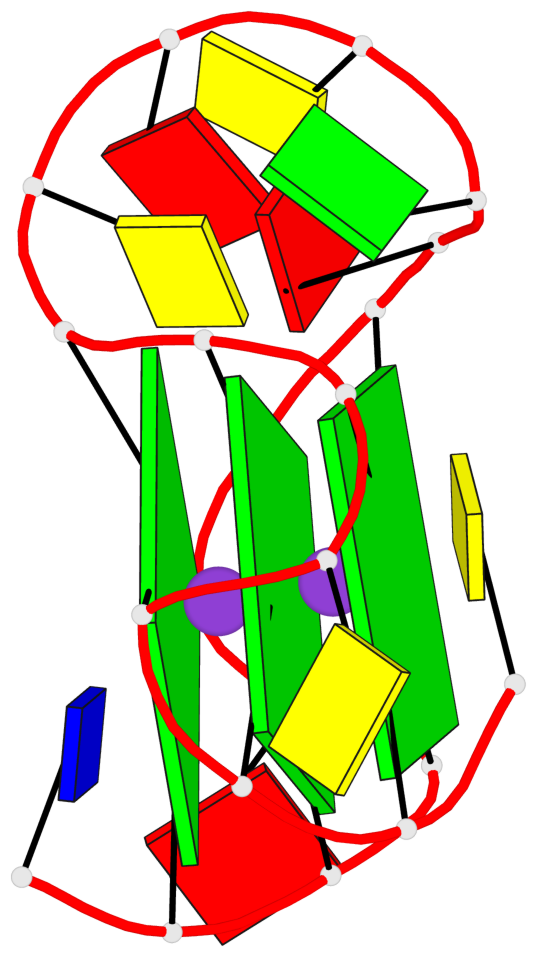
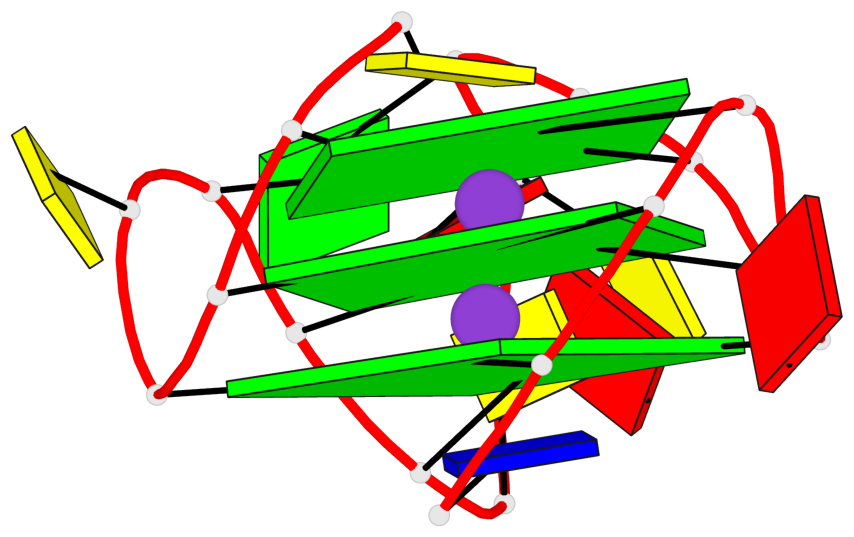
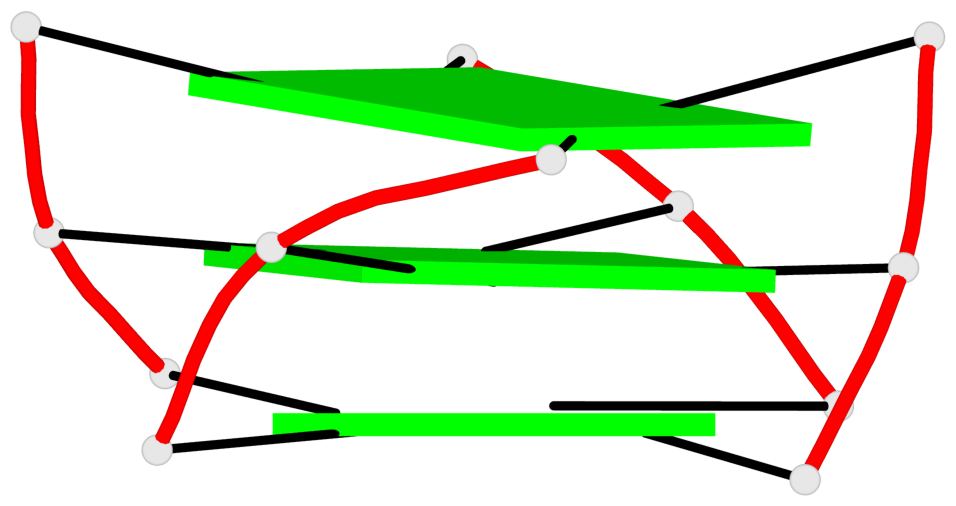
Base-block schematics in six views
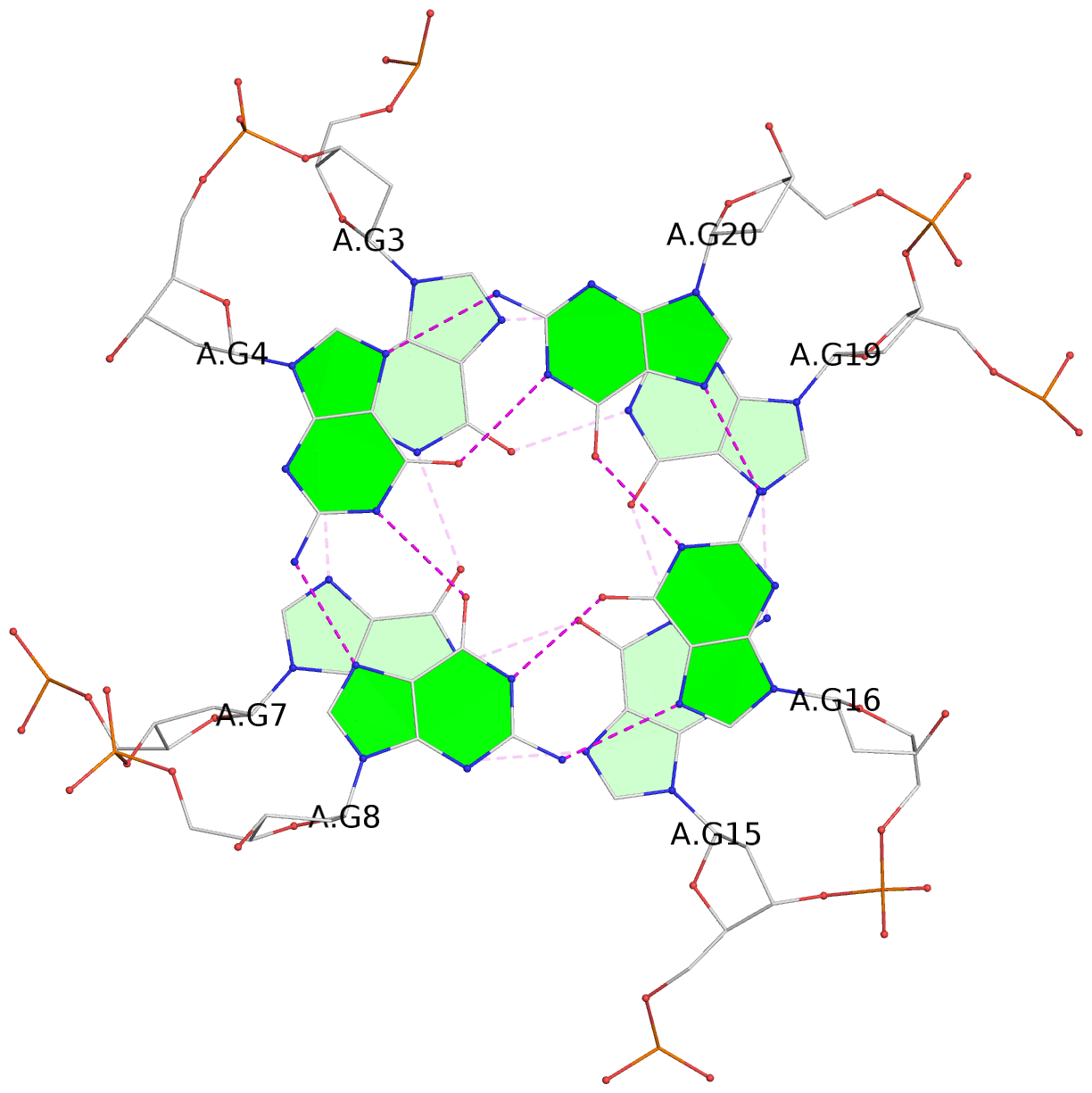
List of 3 G-tetrads
1 glyco-bond=---- sugar=.--- groove=---- planarity=0.276 type=other nts=4 GGGG A.DG2,A.DG6,A.DG14,A.DG18 2 glyco-bond=---- sugar=-.-- groove=---- planarity=0.173 type=other nts=4 GGGG A.DG3,A.DG7,A.DG15,A.DG19 3 glyco-bond=---- sugar=-3-- groove=---- planarity=0.255 type=other nts=4 GGGG A.DG4,A.DG8,A.DG16,A.DG20
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.