Detailed DSSR results for the G-quadruplex: PDB entry 2kvy
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 2kvy
- Class
- DNA
- Method
- NMR
- Summary
- NMR solution structure of the 4:1 complex between an uncharged distamycin a analogue and [d(tggggt)]4
- Reference
- Cosconati S, Marinelli L, Trotta R, Virno A, De Tito S, Romagnoli R, Pagano B, Limongelli V, Giancola C, Baraldi PG, Mayol L, Novellino E, Randazzo A (2010): "Structural and conformational requisites in DNA quadruplex groove binding: another piece to the puzzle." J.Am.Chem.Soc., 132, 6425-6433. doi: 10.1021/ja1003872.
- Abstract
- The study of DNA G-quadruplex stabilizers has enjoyed a great momentum in the late years due to their application as anticancer agents. The recognition of the grooves of these structural motifs is expected to result in a higher degree of selectivity over other DNA structures. Therefore, to achieve an enhanced knowledge on the structural and conformational requisites for quadruplex groove recognition, distamycin A, the only compound for which a pure groove binding has been proven, has been chemically modified. Surprisingly, structural and thermodynamic studies revealed that the absence of Coulombic interactions results in an unprecedented binding position in which both the groove and the 3' end of the DNA are occupied. This further contribution adds another piece to the so far elusive puzzle of the recognition between ligands and DNA quadruplexes and will serve as a platform for a rational design of new groove binders.
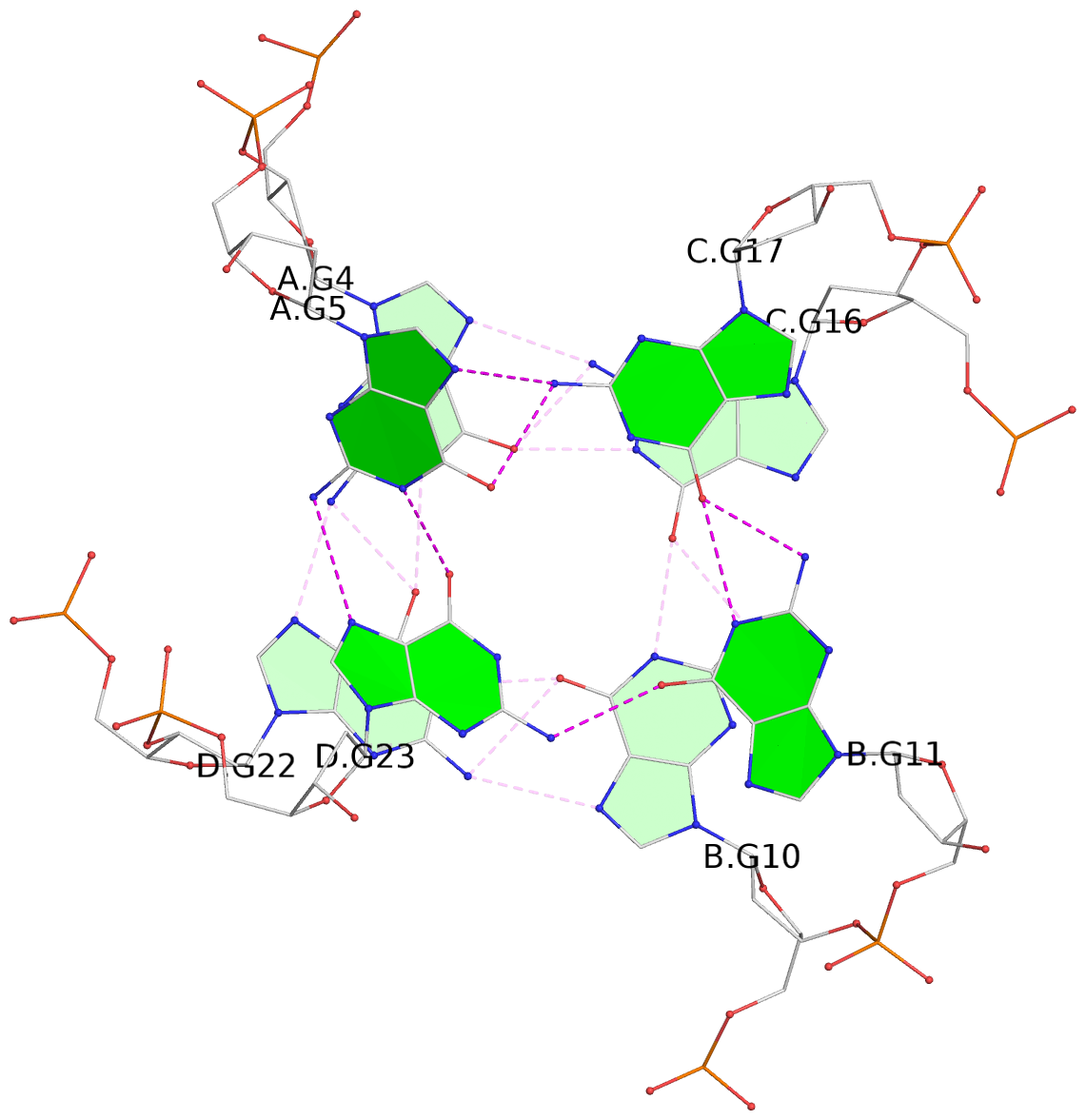
- G4 notes
- 4 G-tetrads, 1 G4 helix, 1 G4 stem, parallel(4+0), UUUU
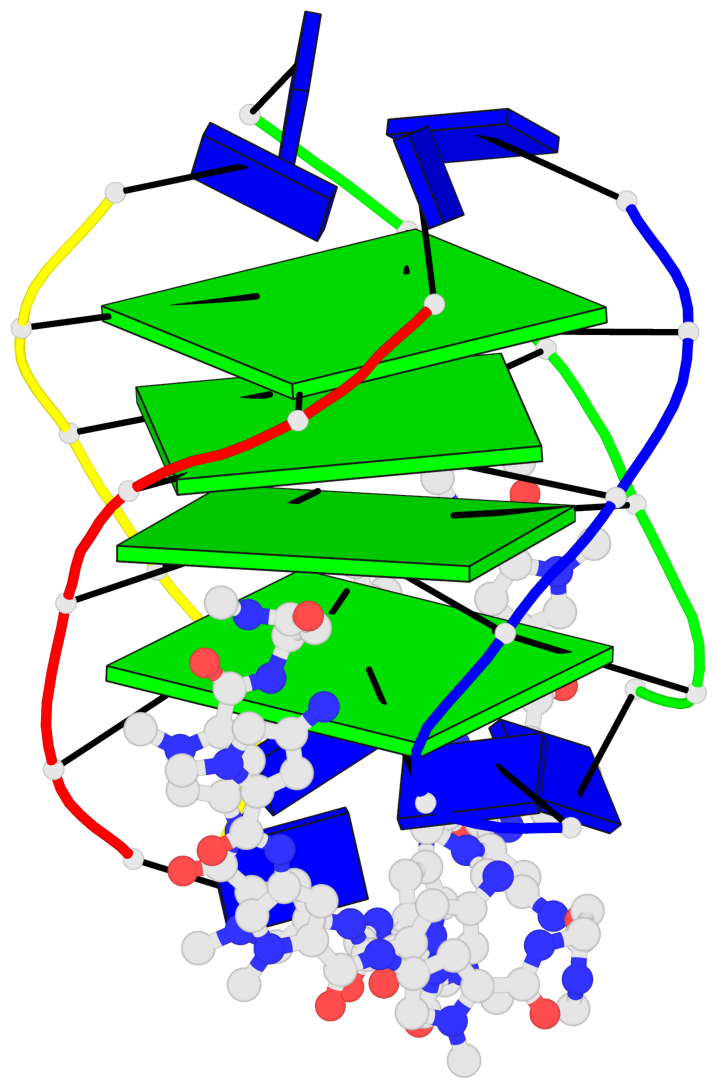
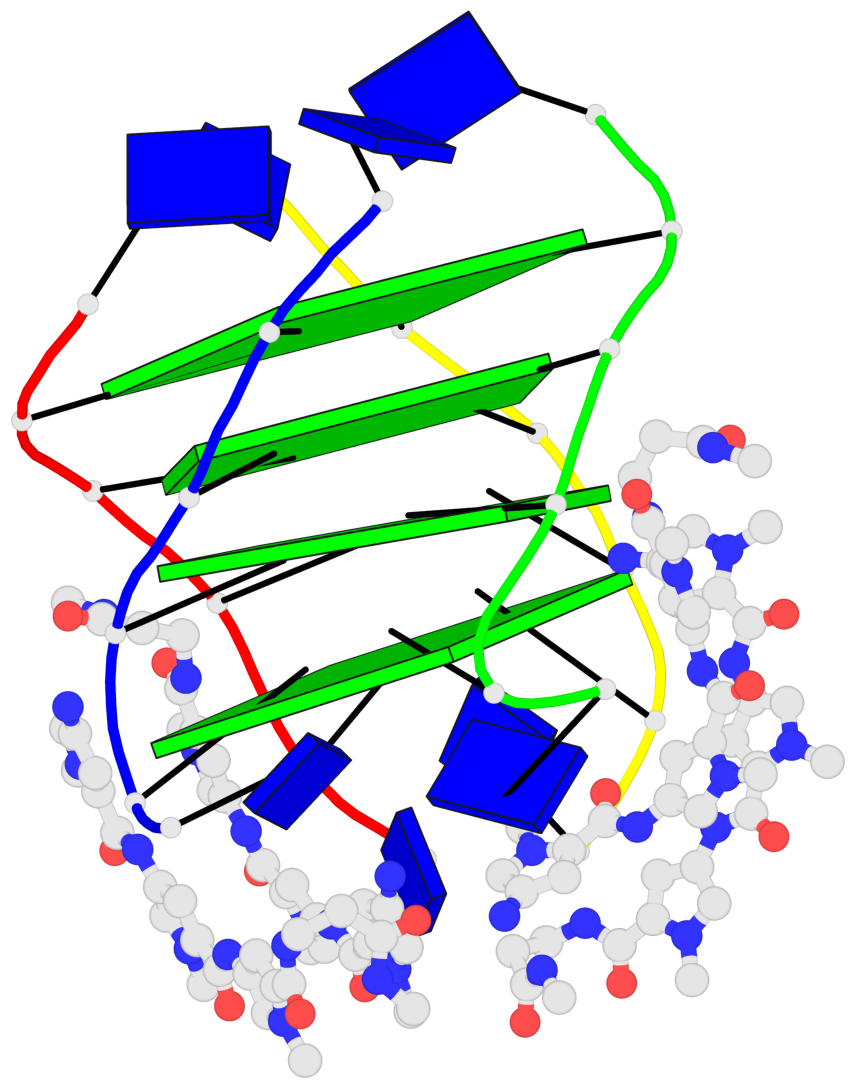
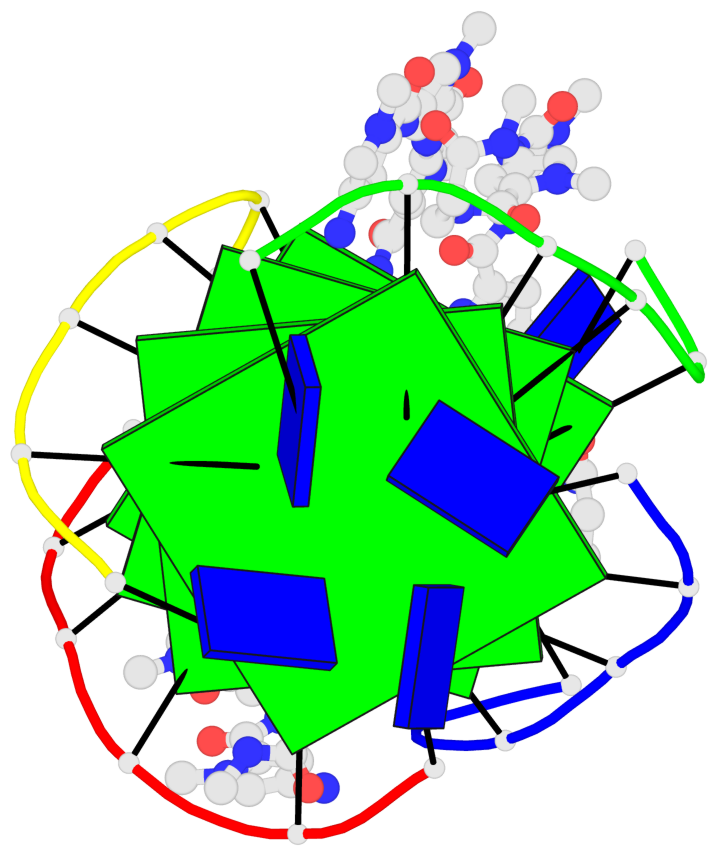
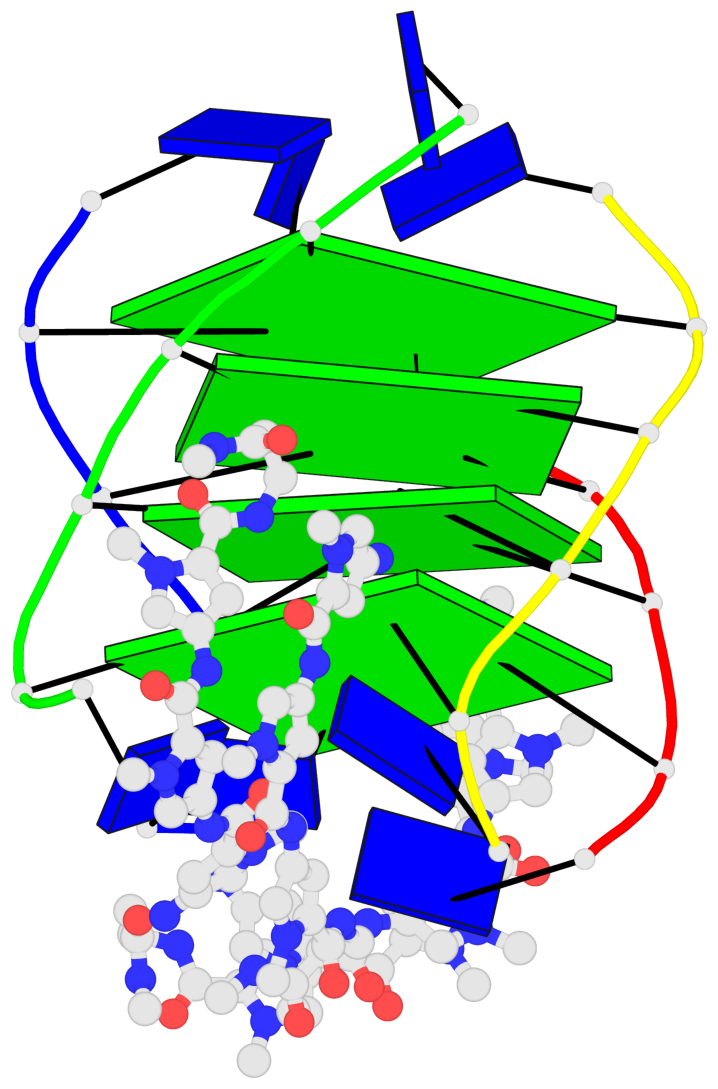
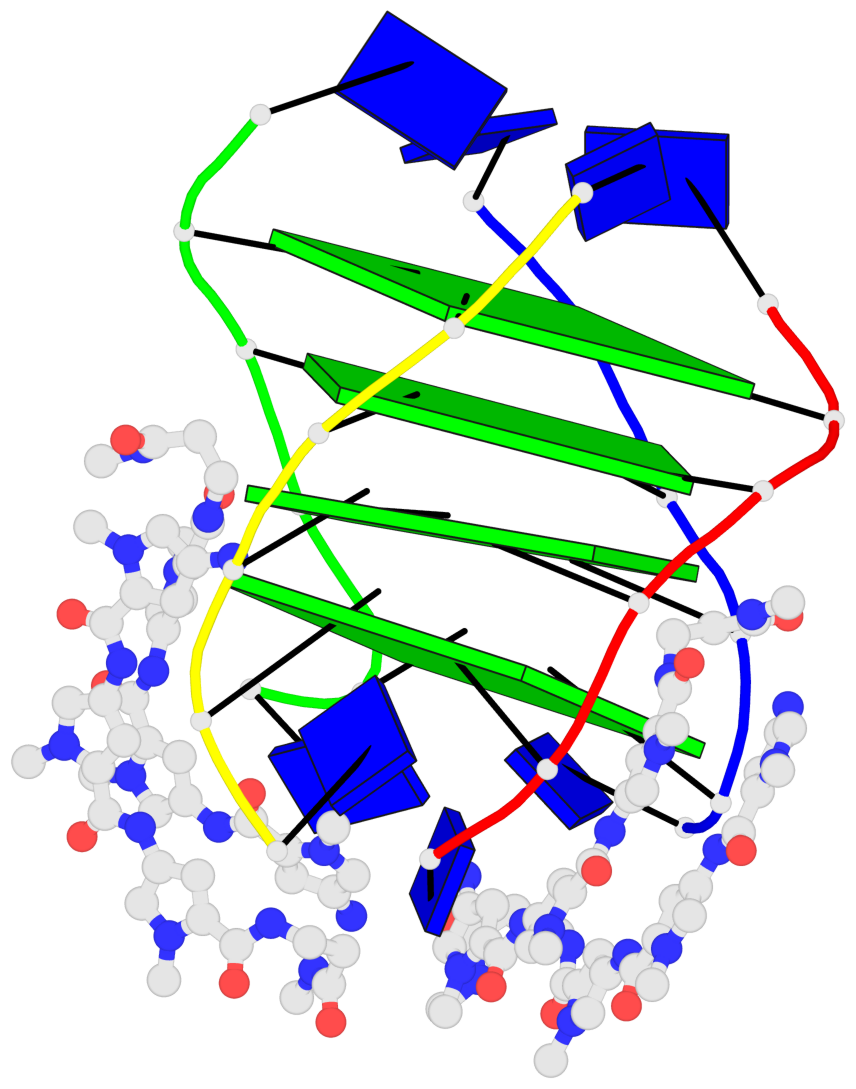
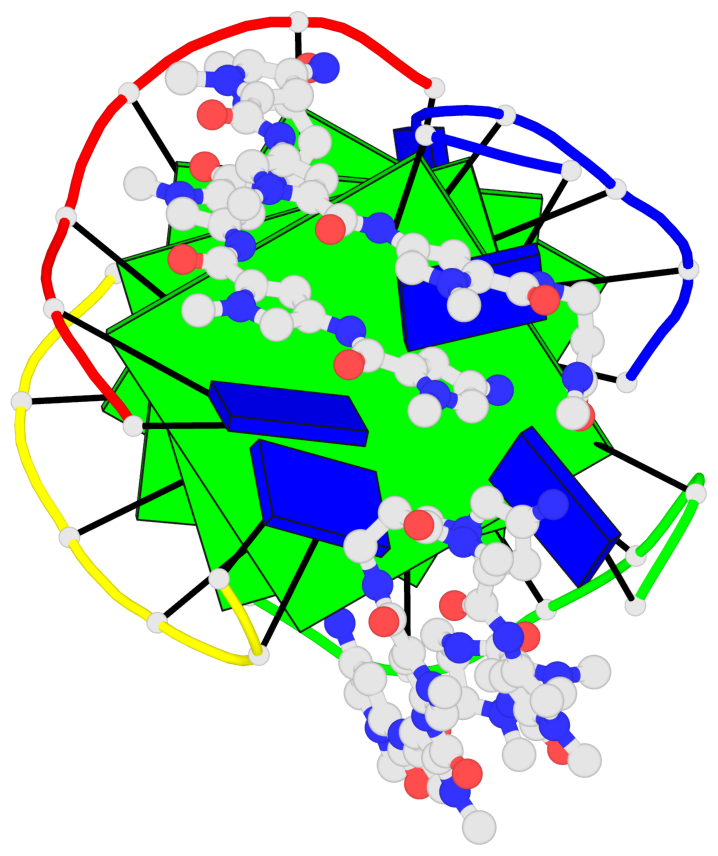
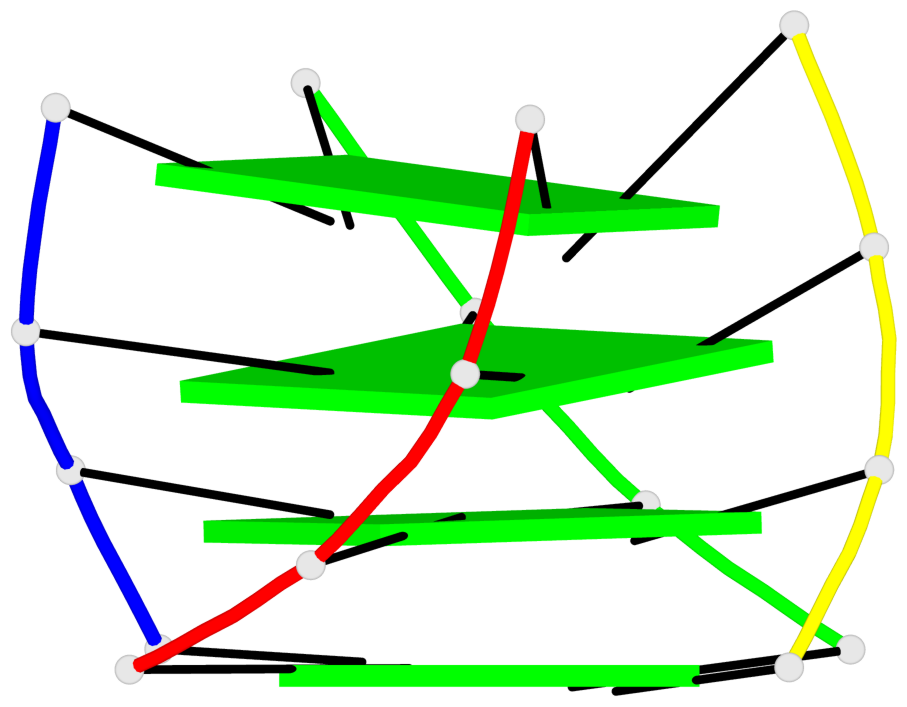
Base-block schematics in six views
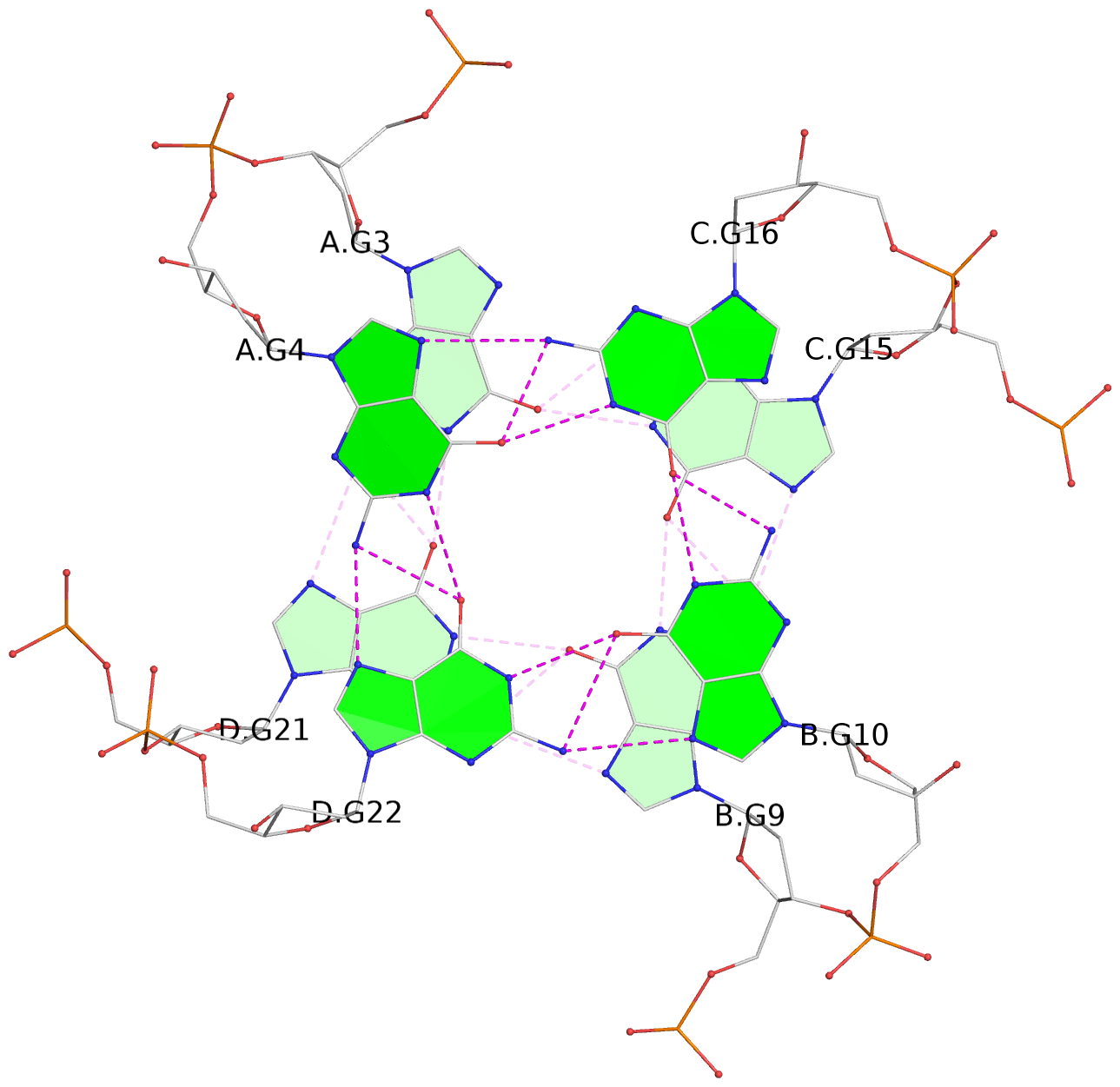
List of 4 G-tetrads
1 glyco-bond=---- sugar=-.-- groove=---- planarity=0.216 type=other nts=4 GGGG A.DG2,D.DG20,B.DG8,C.DG14 2 glyco-bond=---- sugar=-3.. groove=---- planarity=0.226 type=other nts=4 GGGG A.DG3,D.DG21,B.DG9,C.DG15 3 glyco-bond=---- sugar=.3-3 groove=---- planarity=0.379 type=other nts=4 GGGG A.DG4,D.DG22,B.DG10,C.DG16 4 glyco-bond=---- sugar=--.3 groove=---- planarity=0.855 type=bowl nts=4 GGGG A.DG5,D.DG23,B.DG11,C.DG17
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 4 G-tetrad layers, inter-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.