Detailed DSSR results for the G-quadruplex: PDB entry 2l88
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 2l88
- Class
- DNA
- Method
- NMR
- Summary
- Solution structure of all parallel g-quadruplex formed by the oncogene ret promoter sequence
- Reference
- Tong X, Lan W, Zhang X, Wu H, Liu M, Cao C (2011): "Solution structure of all parallel G-quadruplex formed by the oncogene RET promoter sequence." Nucleic Acids Res., 39, 6753-6763. doi: 10.1093/nar/gkr233.
- Abstract
- RET protein functions as a receptor-type tyrosine kinase and has been found to be aberrantly expressed in a wide range of human diseases. A highly GC-rich region upstream of the promoter plays an important role in the transcriptional regulation of RET. Here, we report the NMR solution structure of the major intramolecular G-quadruplex formed on the G-rich strand of this region in K(+) solution. The overall G-quadruplex is composed of three stacked G-tetrad and four syn guanines, which shows distinct features for all parallel-stranded folding topology. The core structure contains one G-tetrad with all syn guanines and two other with all anti-guanines. There are three double-chain reversal loops: the first and the third loops are made of 3 nt G-C-G segments, while the second one contains only 1 nt C10. These loops interact with the core G-tetrads in a specific way that defines and stabilizes the overall G-quadruplex structure and their conformations are in accord with the experimental mutations. The distinct RET promoter G-quadruplex structure suggests that it can be specifically involved in gene regulation and can be an attractive target for pathway-specific drug design.
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(-P-P-P), parallel(4+0), UUUU
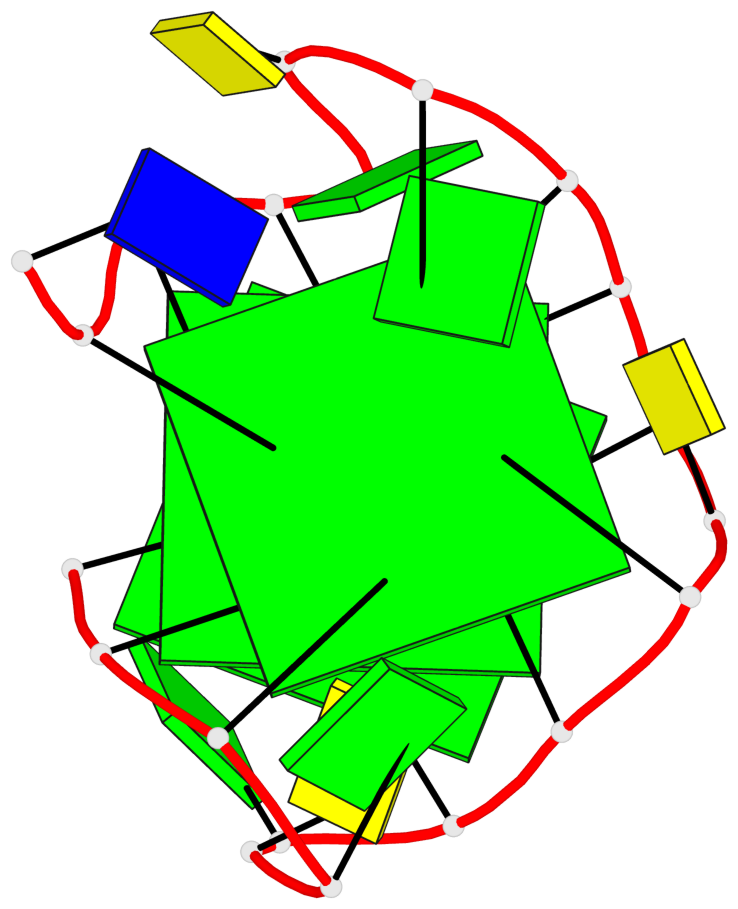
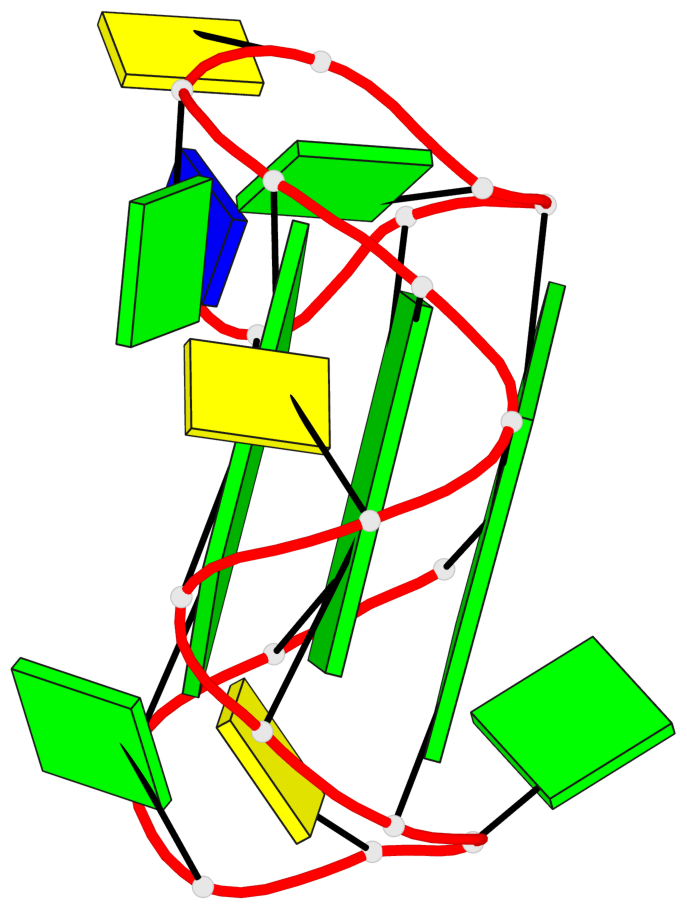
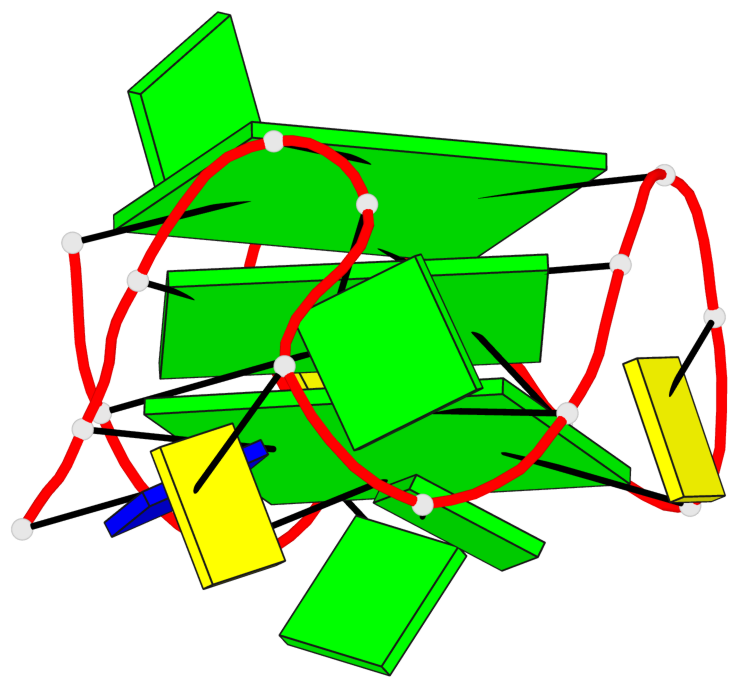
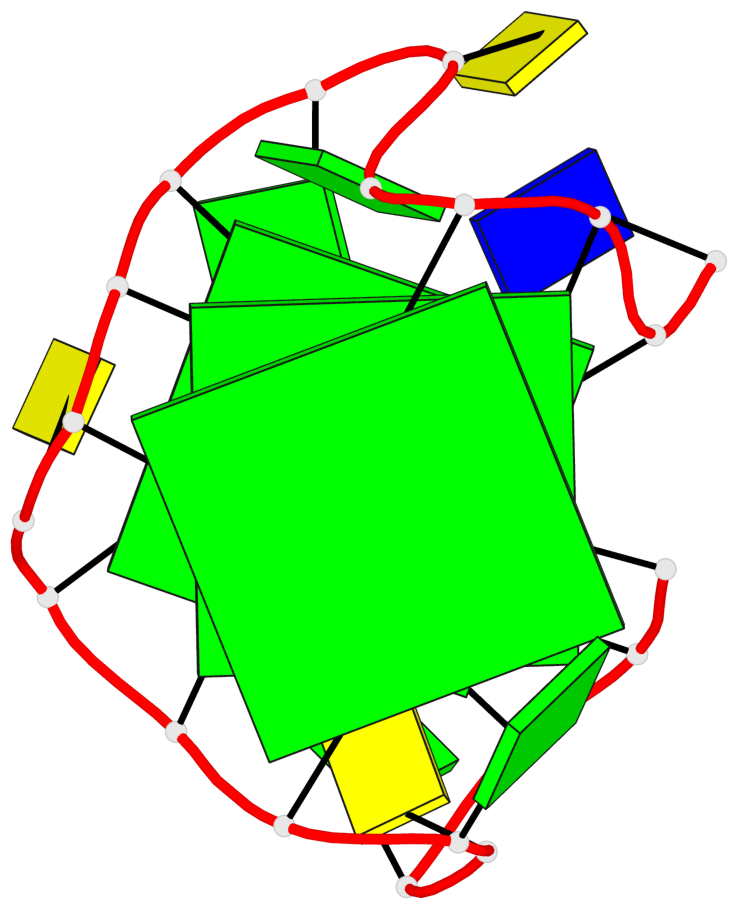
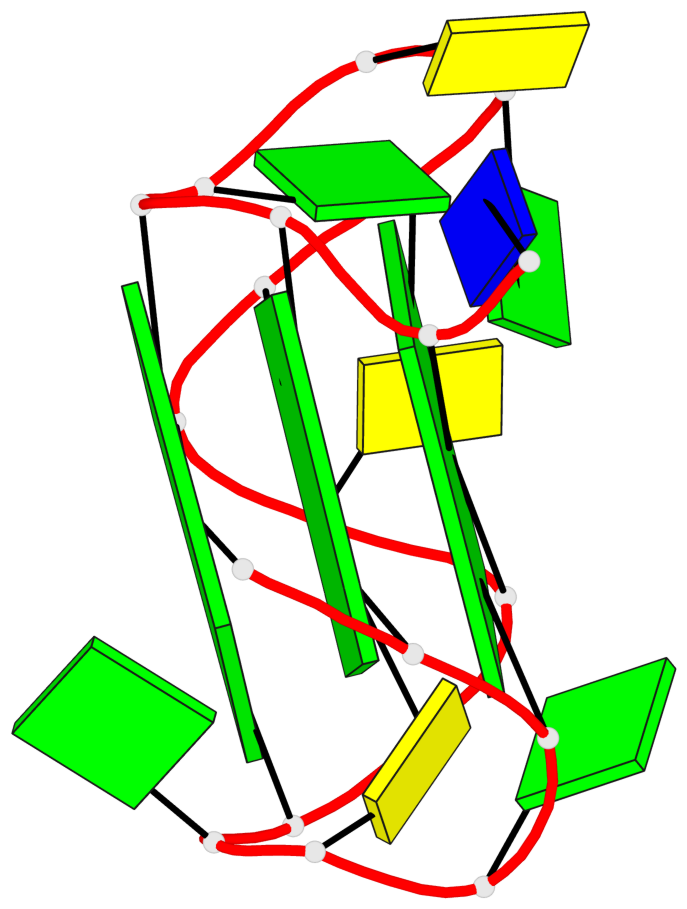
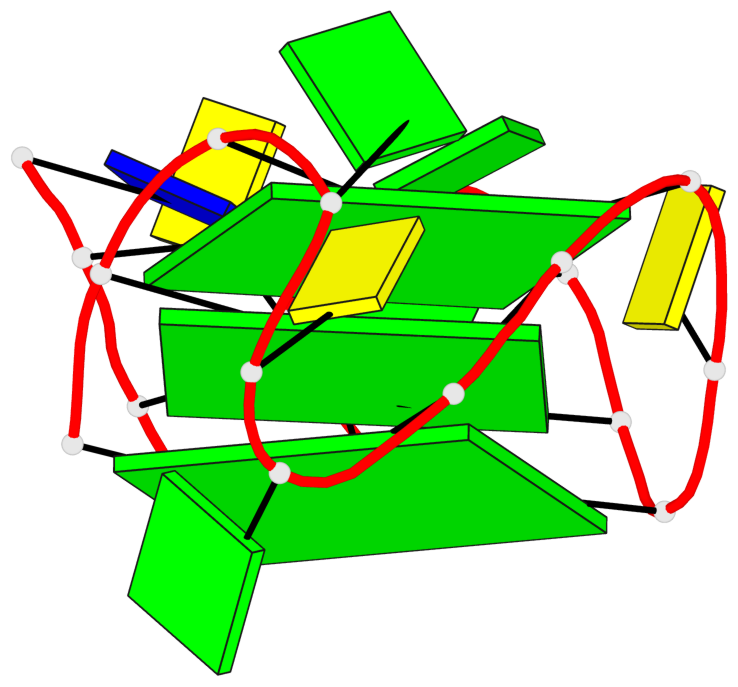
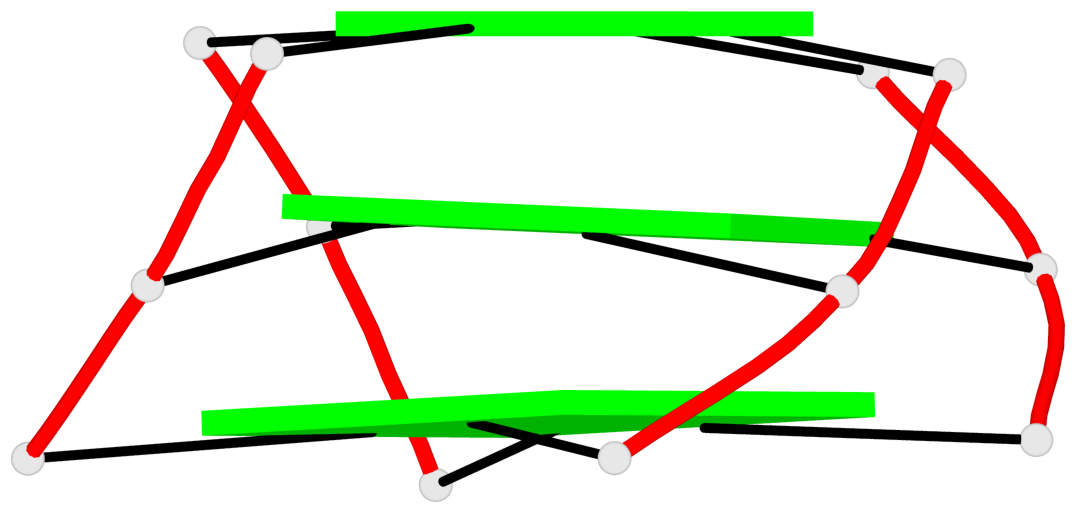
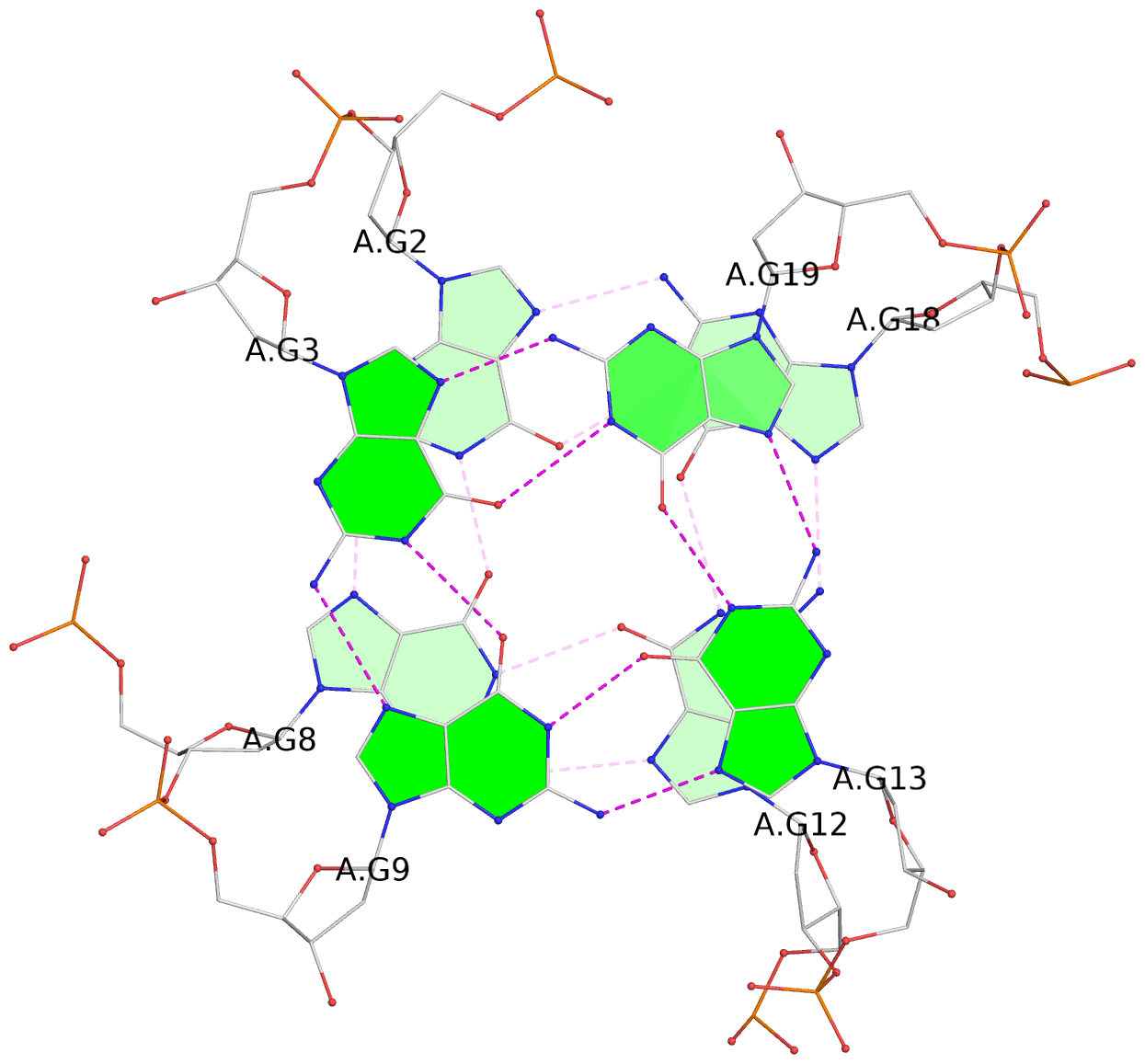
Base-block schematics in six views
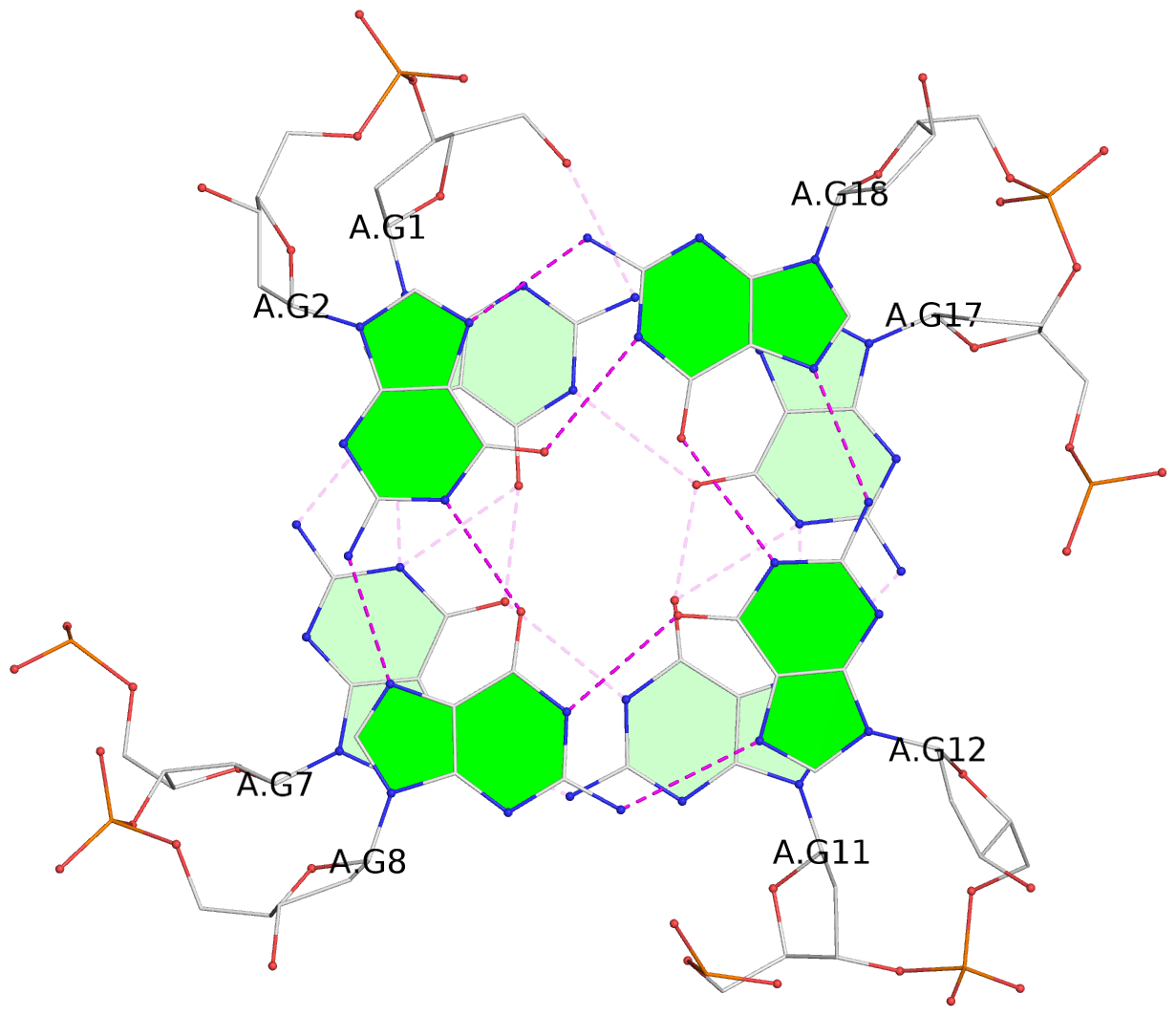
List of 3 G-tetrads
1 glyco-bond=ssss sugar=3.-. groove=---- planarity=0.121 type=planar nts=4 GGGG A.DG1,A.DG7,A.DG11,A.DG17 2 glyco-bond=---- sugar=3.3. groove=---- planarity=0.136 type=planar nts=4 GGGG A.DG2,A.DG8,A.DG12,A.DG18 3 glyco-bond=---- sugar=--3. groove=---- planarity=0.256 type=other nts=4 GGGG A.DG3,A.DG9,A.DG13,A.DG19
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.