Detailed DSSR results for the G-quadruplex: PDB entry 2la5
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 2la5
- Class
- RNA binding protein-RNA
- Method
- NMR
- Summary
- RNA duplex-quadruplex junction complex with fmrp rgg peptide
- Reference
- Phan AT, Kuryavyi V, Darnell JC, Serganov A, Majumdar A, Ilin S, Raslin T, Polonskaia A, Chen C, Clain D, Darnell RB, Patel DJ (2011): "Structure-function studies of FMRP RGG peptide recognition of an RNA duplex-quadruplex junction." Nat.Struct.Mol.Biol., 18, 796-804. doi: 10.1038/nsmb.2064.
- Abstract
- We have determined the solution structure of the complex between an arginine-glycine-rich RGG peptide from the human fragile X mental retardation protein (FMRP) and an in vitro-selected guanine-rich (G-rich) sc1 RNA. The bound RNA forms a newly discovered G-quadruplex separated from the flanking duplex stem by a mixed junctional tetrad. The RGG peptide is positioned along the major groove of the RNA duplex, with the G-quadruplex forcing a sharp turn of R(10)GGGGR(15) at the duplex-quadruplex junction. Arg10 and Arg15 form cross-strand specificity-determining intermolecular hydrogen bonds with the major-groove edges of guanines of adjacent Watson-Crick G•C pairs. Filter-binding assays on RNA and peptide mutations identify and validate contributions of peptide-RNA intermolecular contacts and shape complementarity to molecular recognition. These findings on FMRP RGG domain recognition by a combination of G-quadruplex and surrounding RNA sequences have implications for the recognition of other genomic G-rich RNAs.
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 2(-P-P-P), parallel(4+0), UUUU
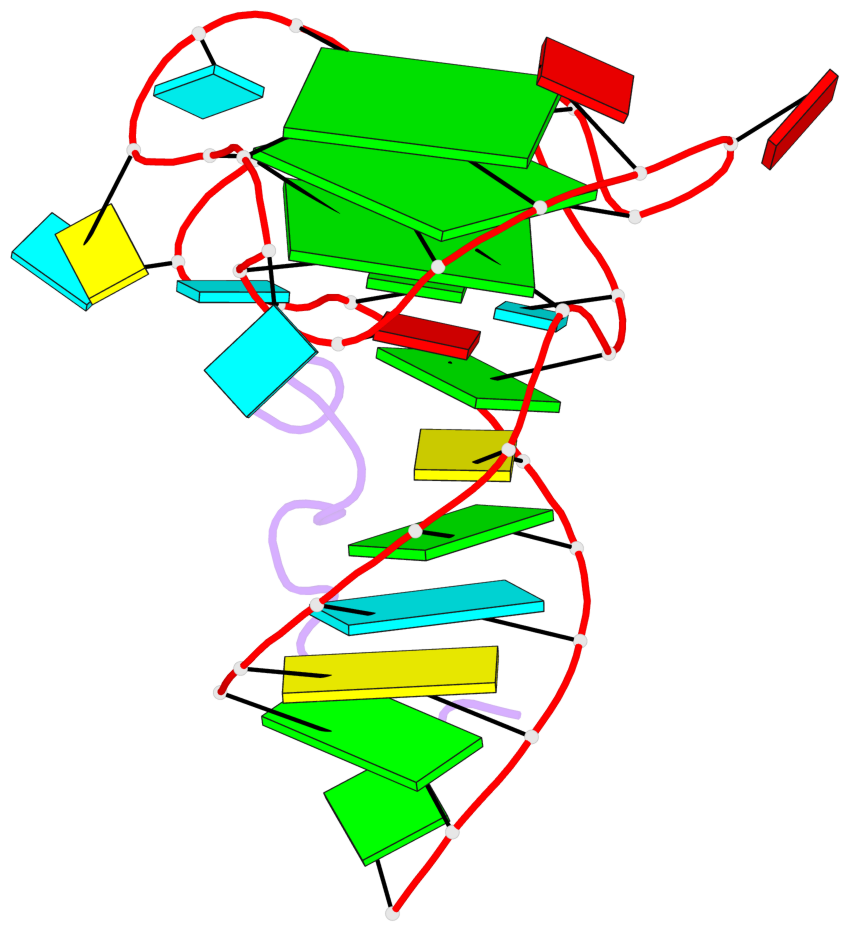
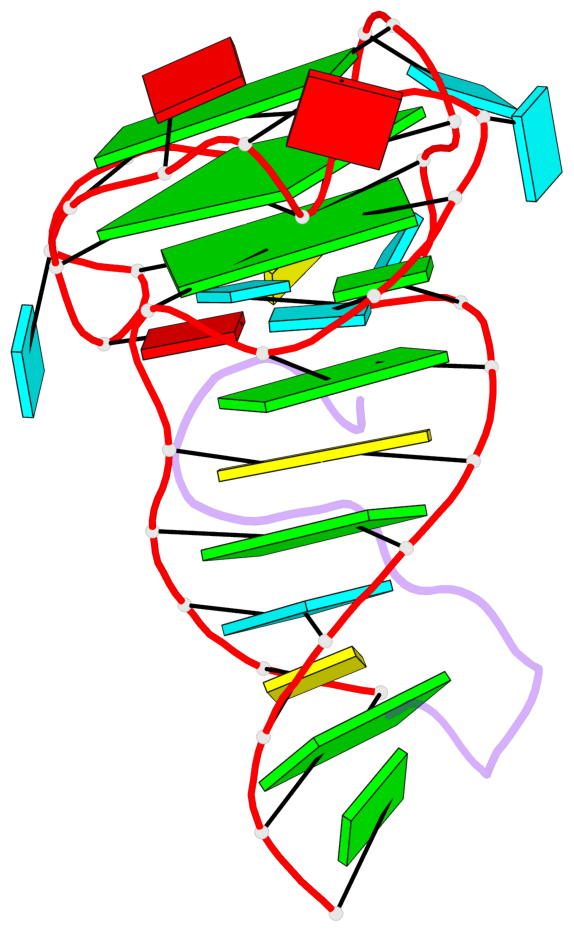
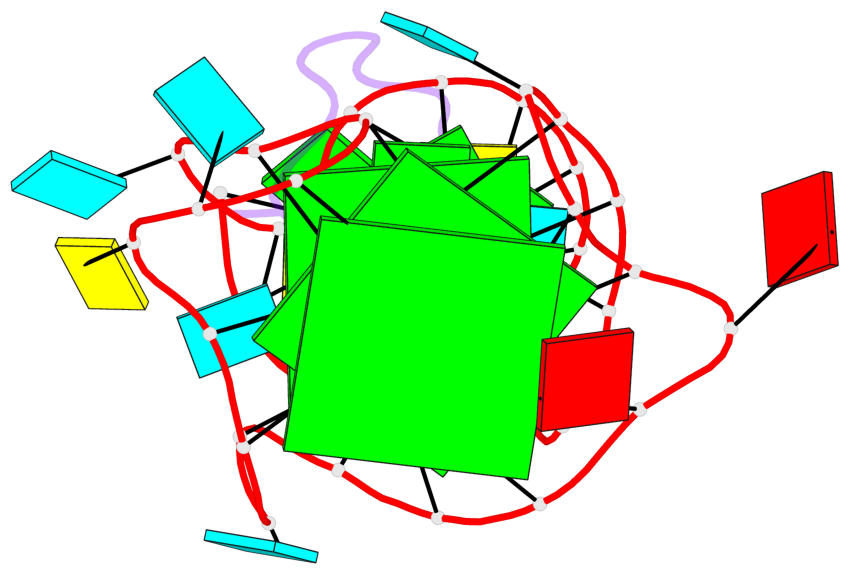
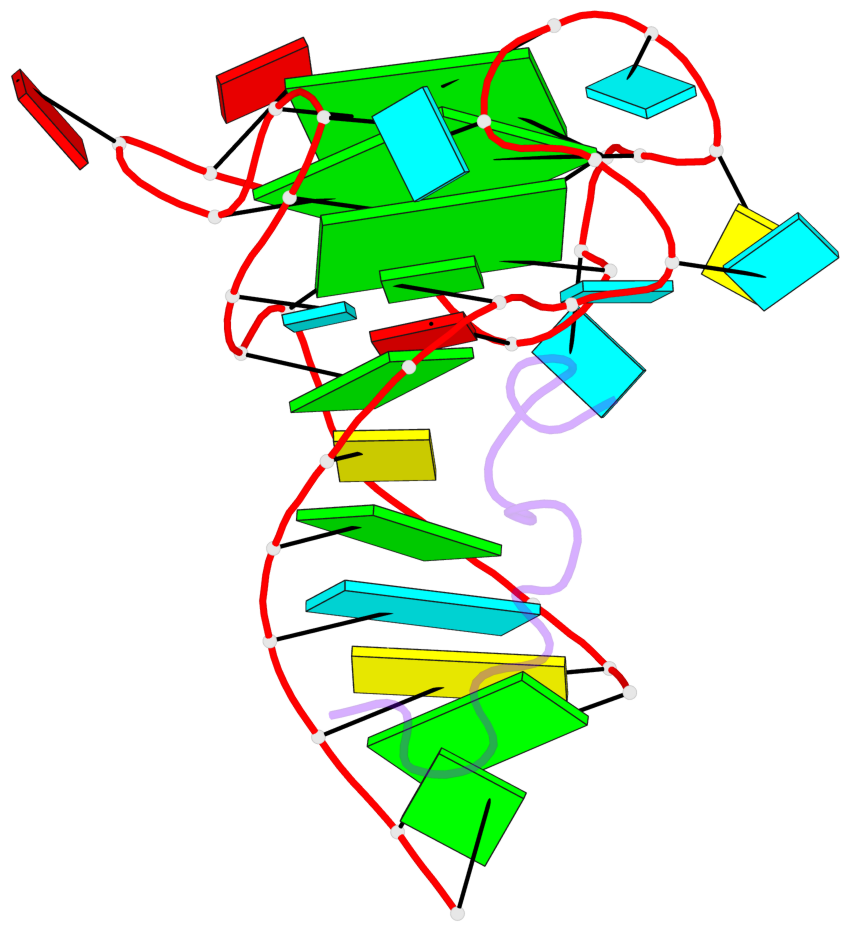
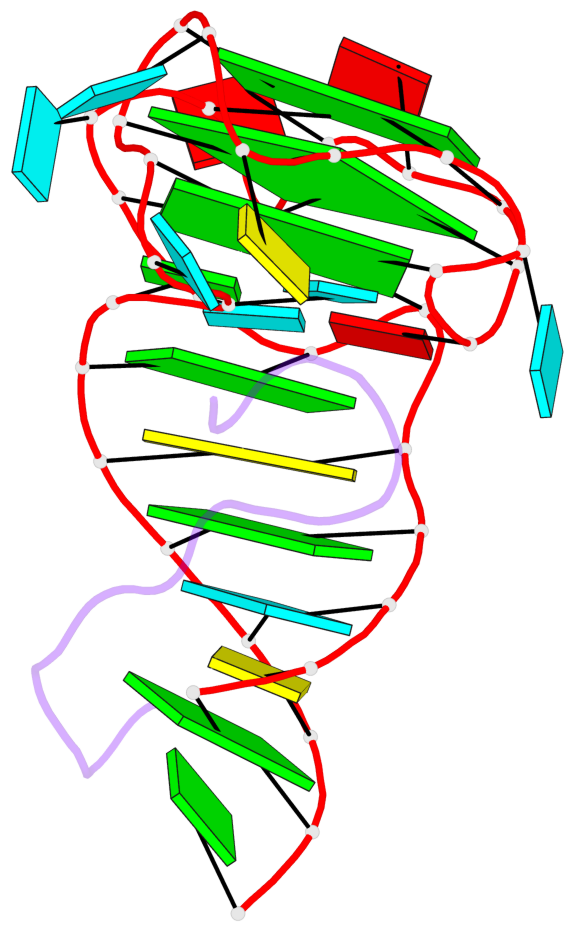
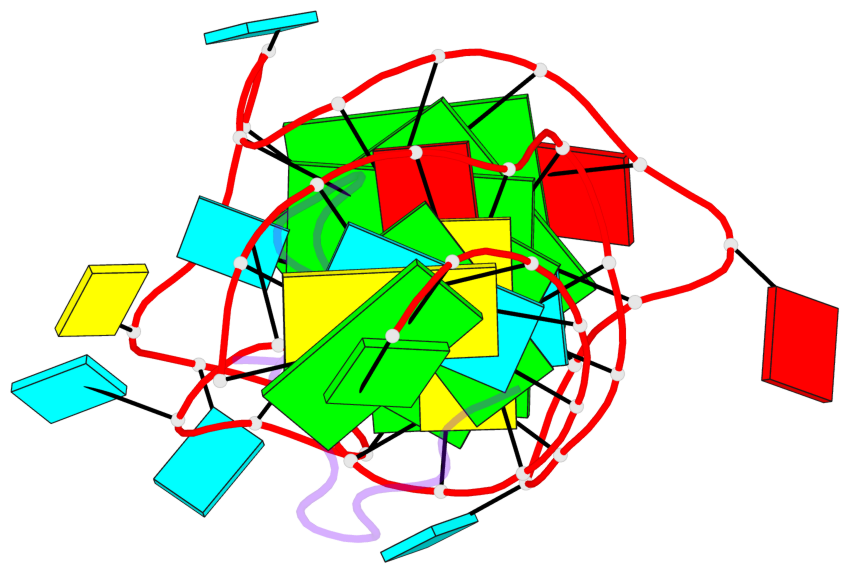
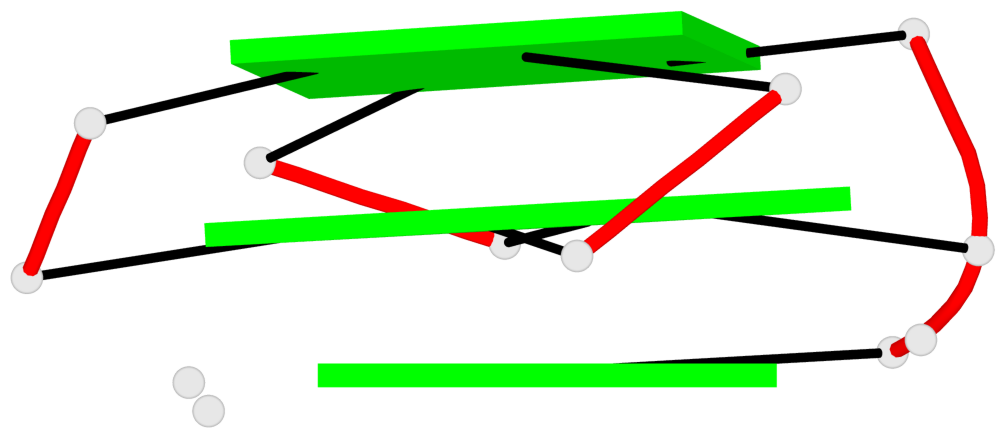
Base-block schematics in six views
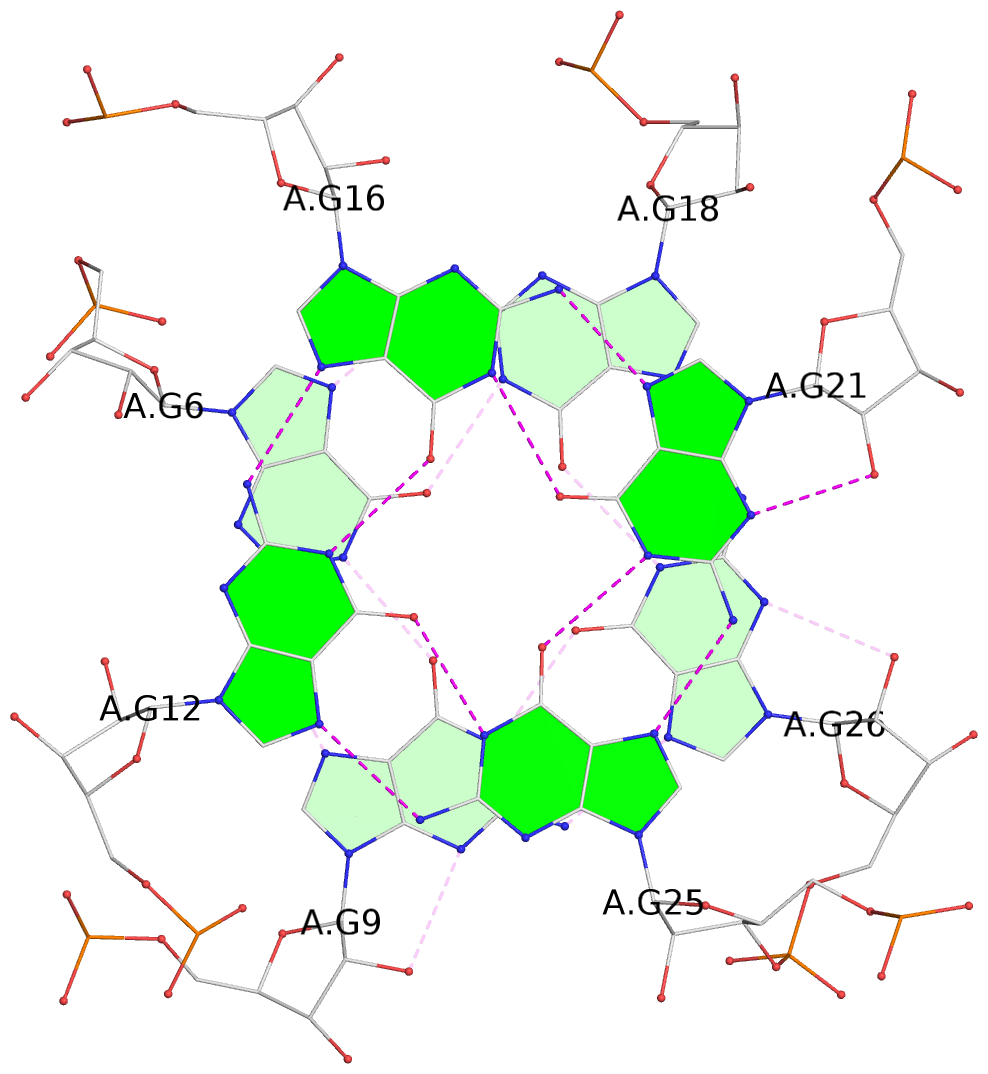
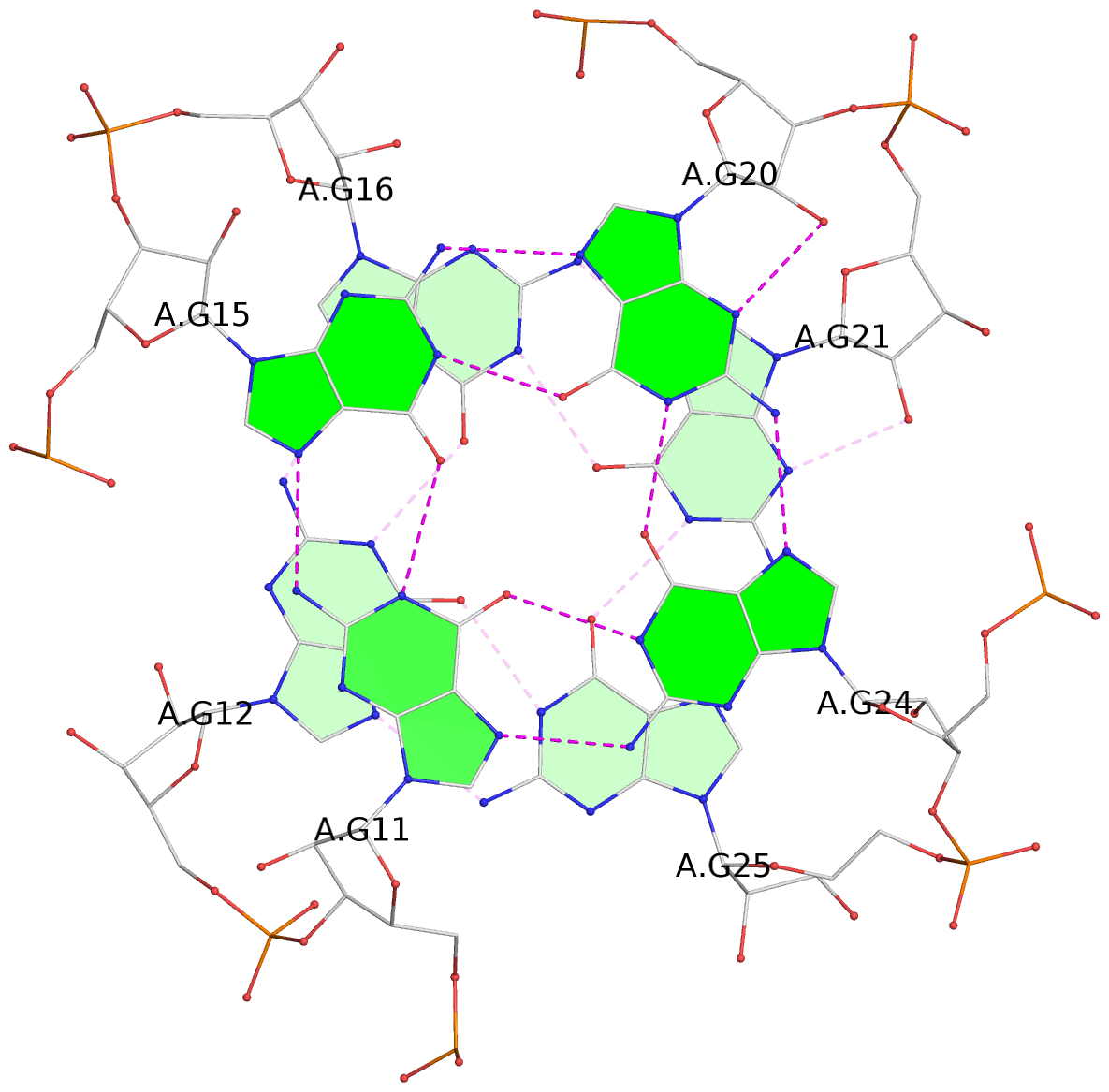
List of 3 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.176 type=other nts=4 GGGG A.G6,A.G9,A.G26,A.G18 2 glyco-bond=---- sugar=3--- groove=---- planarity=0.195 type=other nts=4 GGGG A.G11,A.G15,A.G20,A.G24 3 glyco-bond=---- sugar=---3 groove=---- planarity=0.106 type=planar nts=4 GGGG A.G12,A.G16,A.G21,A.G25
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 2 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 2(-P-P-P), parallel(4+0)
List of 1 non-stem G4-loop (including the two closing Gs)
1 type=lateral helix=#1 nts=4 GGUG A.G6,A.G7,A.U8,A.G9