Detailed DSSR results for the G-quadruplex: PDB entry 2lod
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 2lod
- Class
- DNA
- Method
- NMR
- Summary
- Solution-state structure of an intramolecular g-quadruplex with propeller, diagonal and edgewise loops
- Reference
- Marusic M, Sket P, Bauer L, Viglasky V, Plavec J (2012): "Solution-state structure of an intramolecular G-quadruplex with propeller, diagonal and edgewise loops." Nucleic Acids Res., 40, 6946-6956. doi: 10.1093/nar/gks329.
- Abstract
- We herein report on the formation and high-resolution NMR solution-state structure determination of a G-quadruplex adopted by d[G(3)ATG(3)ACACAG(4)ACG(3)] comprised of four G-tracts with the third one consisting of four guanines that are intervened with non-G streches of different lengths. A single intramolecular antiparallel (3+1) G-quadruplex exhibits three stacked G-quartets connected with propeller, diagonal and edgewise loops of different lengths. The propeller and edgewise loops are well structured, whereas the longer diagonal loop is more flexible. To the best of our knowledge, this is the first high-resolution G-quadruplex structure where all of the three main loop types are present.
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(-PD+Lw), U3D(3+1), UUUD
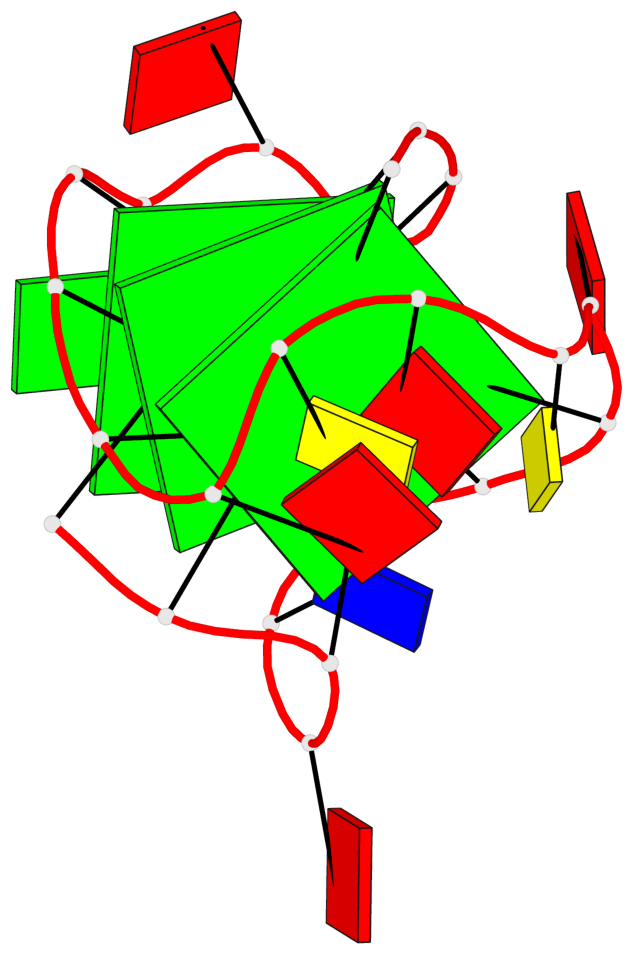
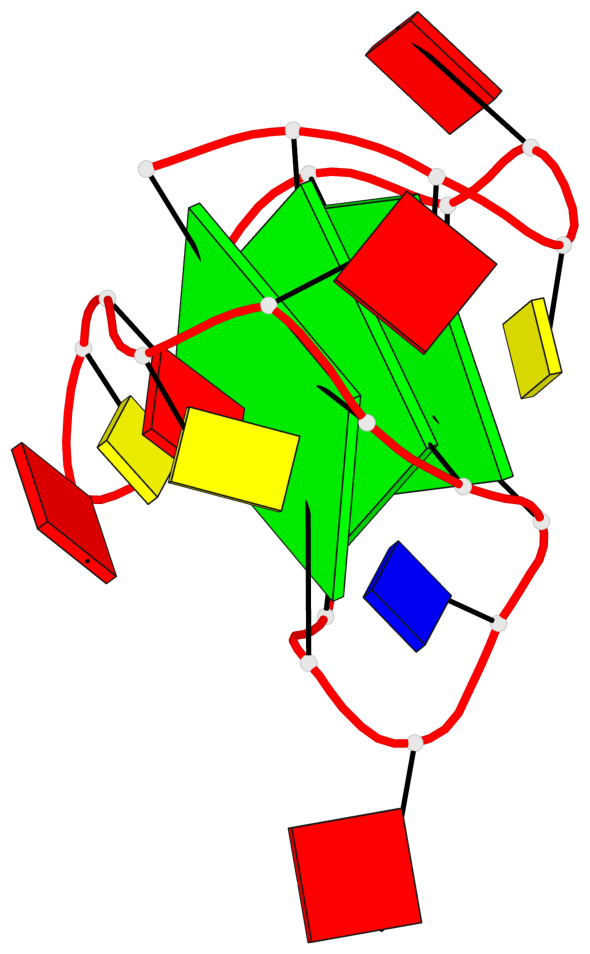
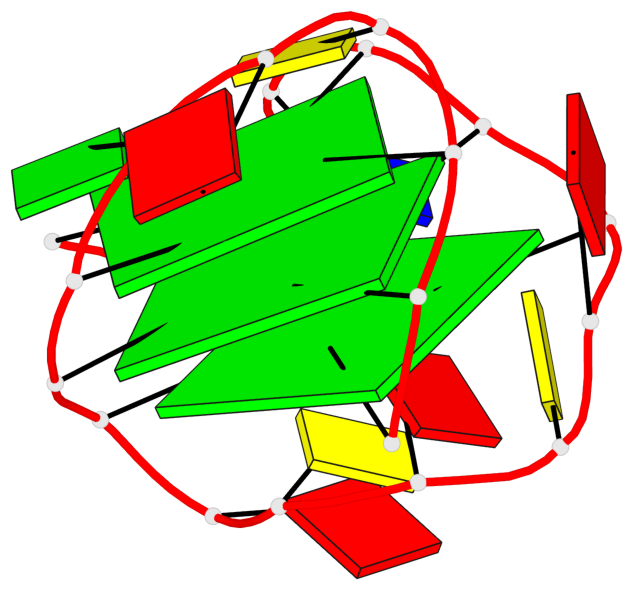
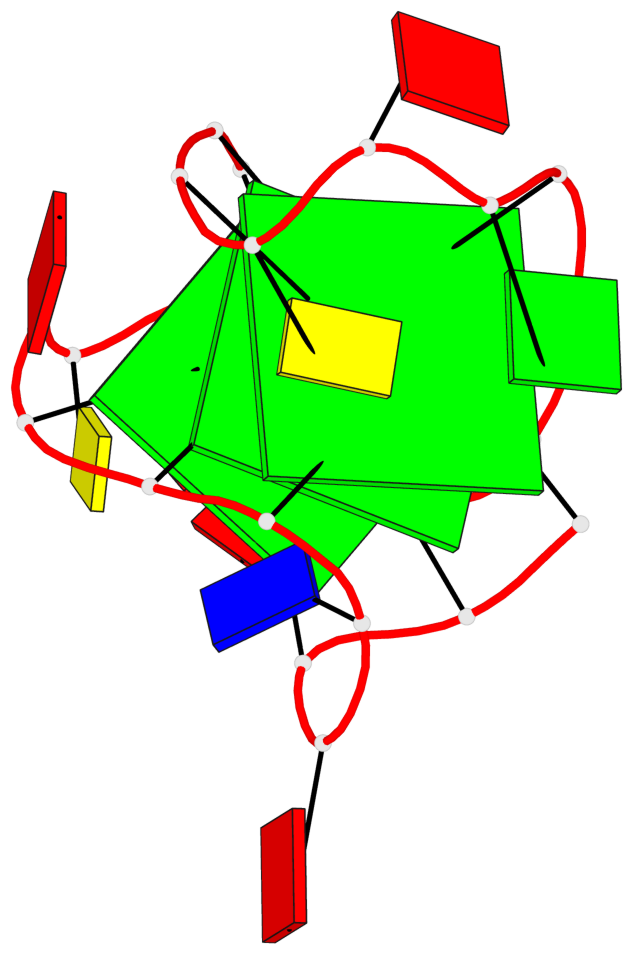
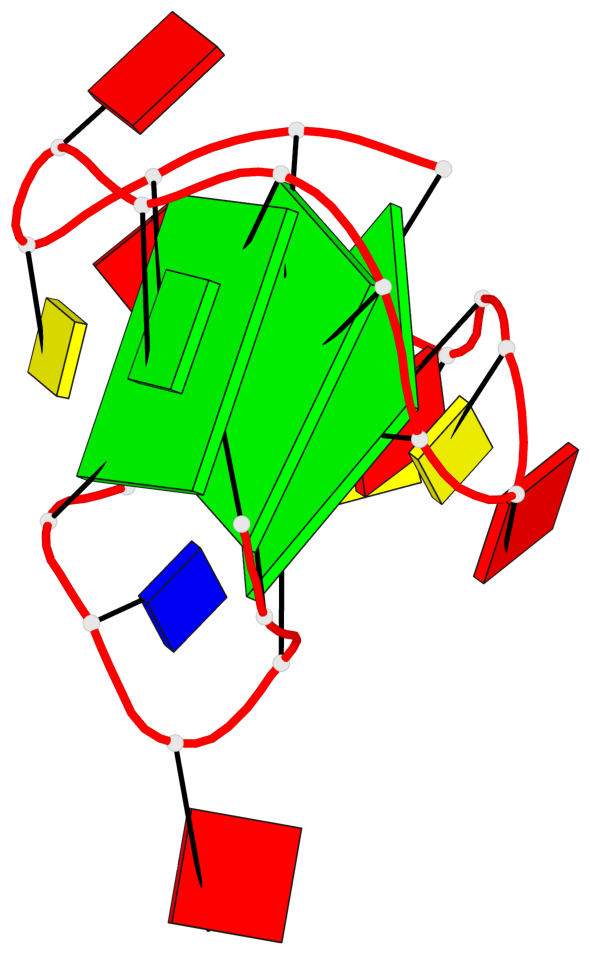
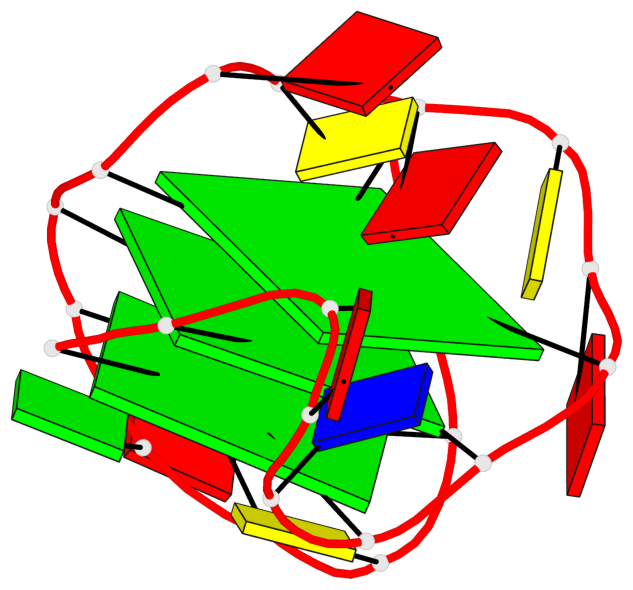
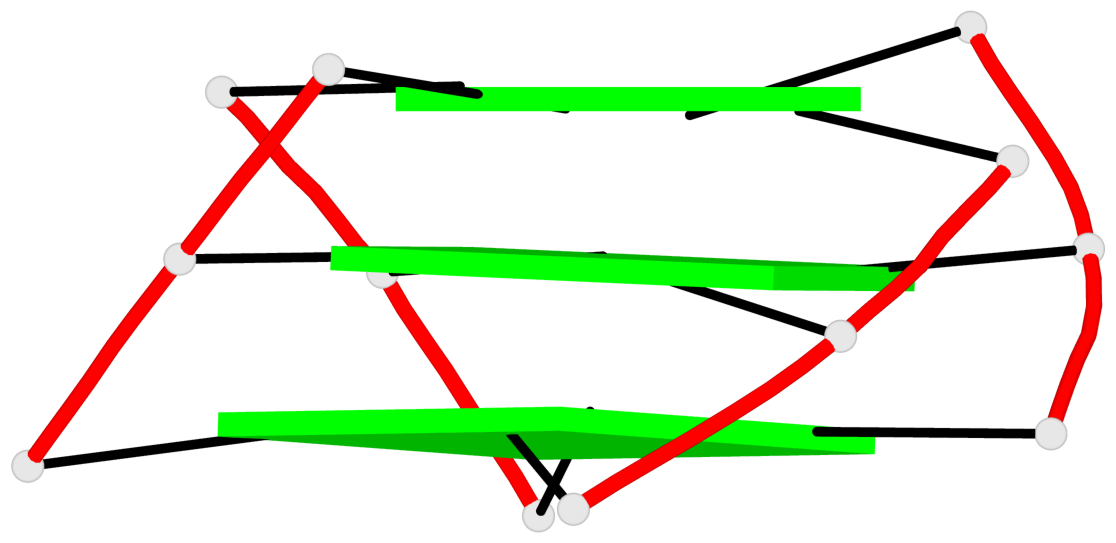
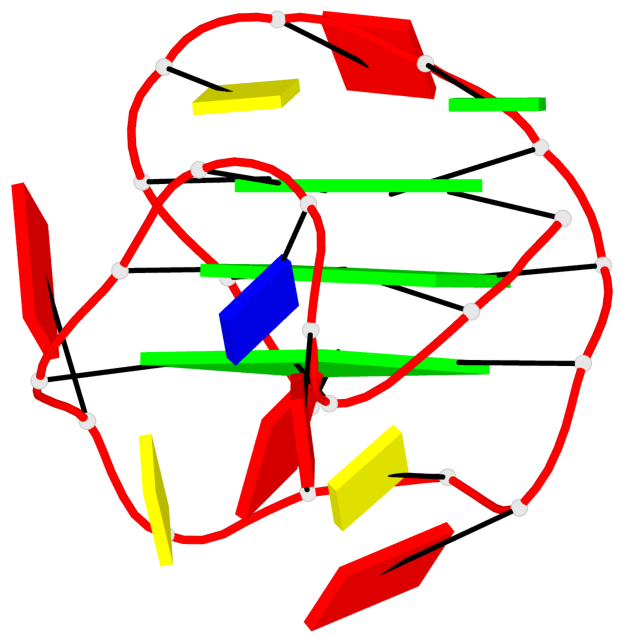
Base-block schematics in six views
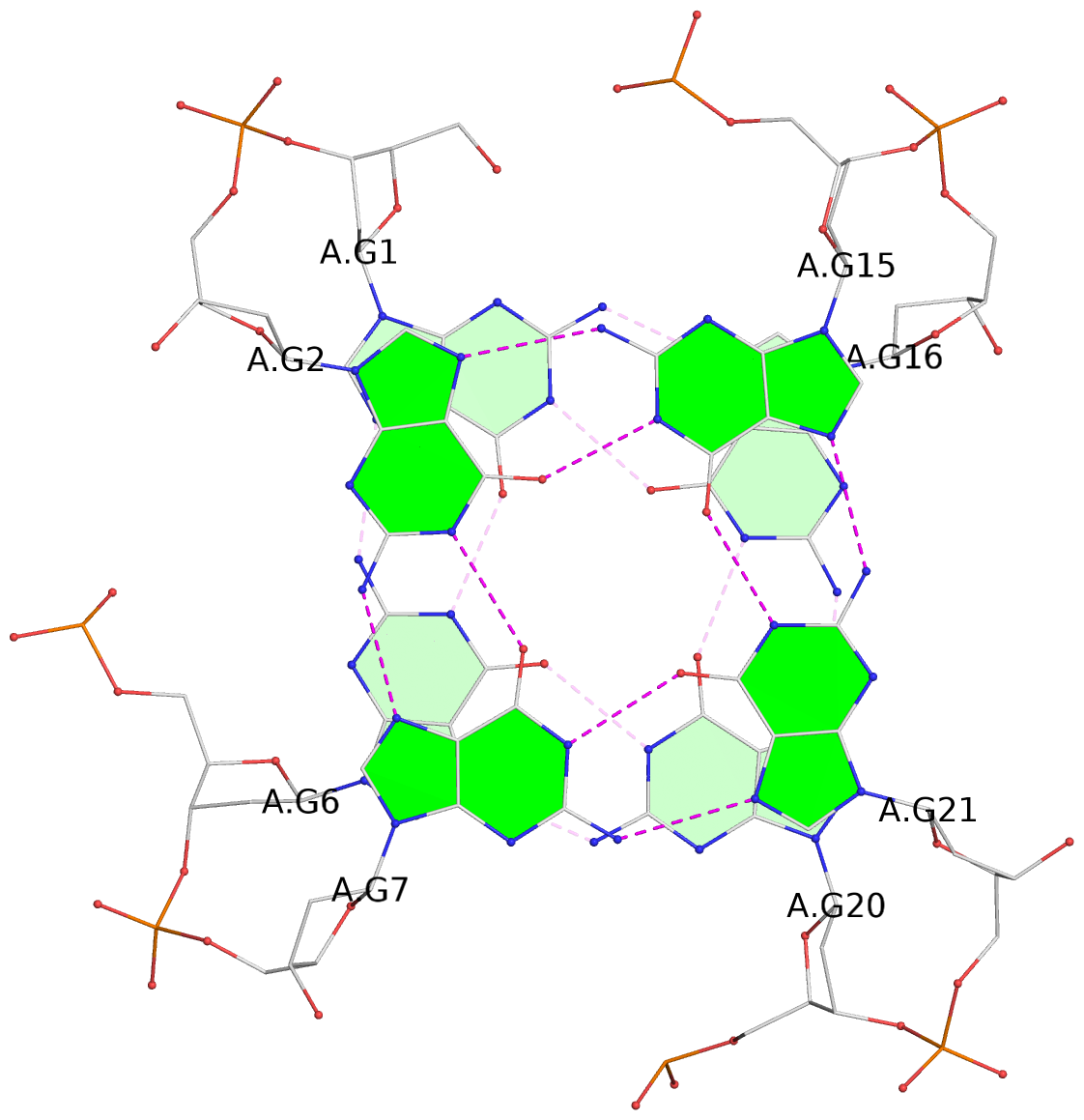
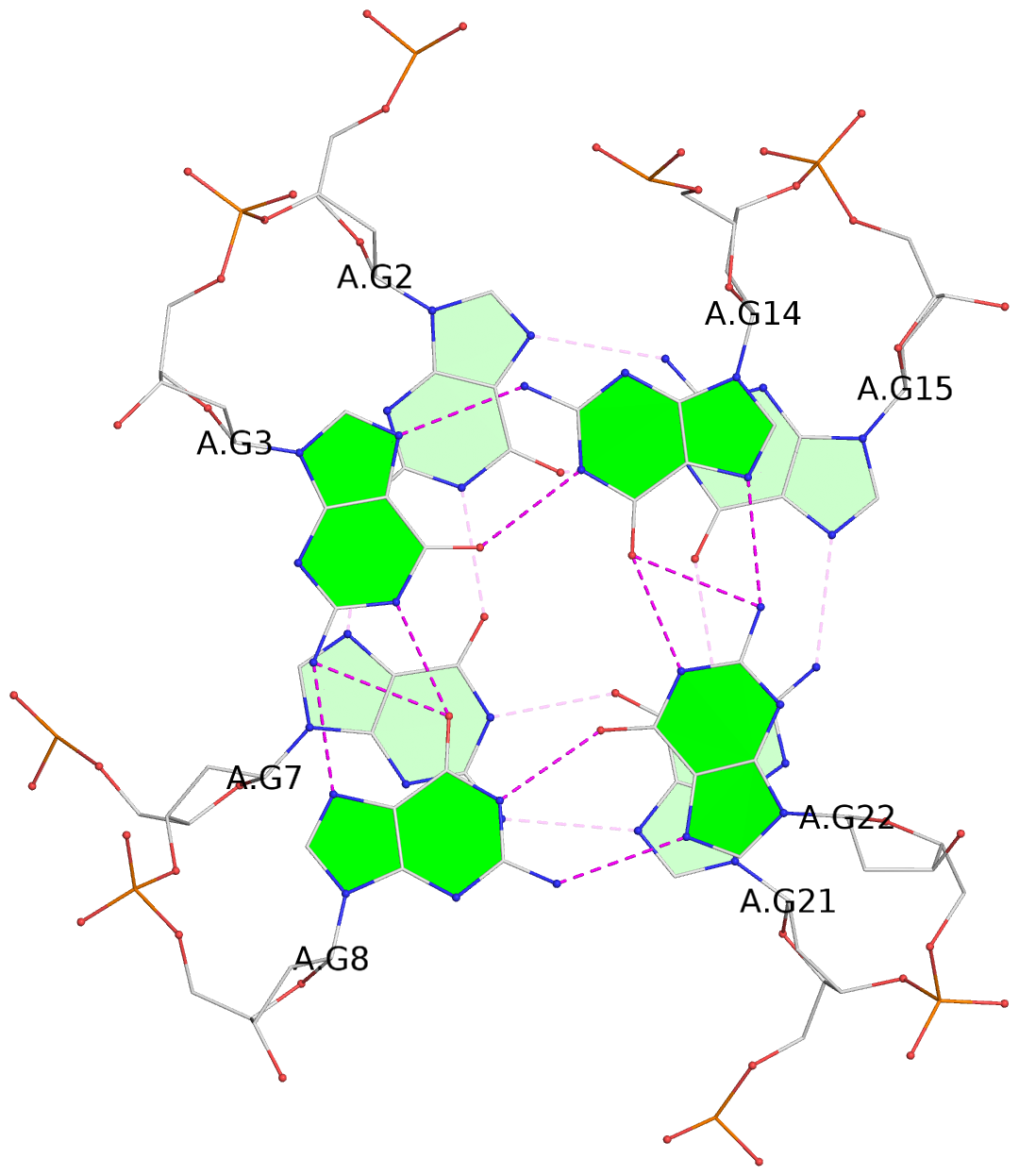
List of 3 G-tetrads
1 glyco-bond=sss- sugar=---- groove=--wn planarity=0.219 type=other nts=4 GGGG A.DG1,A.DG6,A.DG20,A.DG16 2 glyco-bond=---s sugar=---- groove=--wn planarity=0.109 type=planar nts=4 GGGG A.DG2,A.DG7,A.DG21,A.DG15 3 glyco-bond=---s sugar=---- groove=--wn planarity=0.309 type=bowl nts=4 GGGG A.DG3,A.DG8,A.DG22,A.DG14
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.