Detailed DSSR results for the G-quadruplex: PDB entry 2lpw
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 2lpw
- Class
- DNA
- Method
- NMR
- Summary
- Human ceb25 minisatellite g-quadruplex
- Reference
- Amrane S, Adrian M, Heddi B, Serero A, Nicolas A, Mergny JL, Phan AT (2012): "Formation of Pearl-Necklace Monomorphic G-Quadruplexes in the Human CEB25 Minisatellite." J.Am.Chem.Soc., 134, 5807-5816. doi: 10.1021/ja208993r.
- Abstract
- CEB25 is a human minisatellite locus, composed of slightly polymorphic 52-nucleotide (nt) tandem repeats. Genetically, most if not all individuals of the human population are heterozygous, carrying alleles ranging from 0.5 to 20 kb, maintained by mendelian inheritance but also subject to germline instability. To provide insights on the biological role of CEB25, we interrogated its structural features. We report the NMR structure of the G-quadruplex formed by the conserved 26-nt G-rich fragment of the CEB25 motif. In K(+) solution, this sequence forms a propeller-type parallel-stranded G-quadruplex involving a 9-nt central double-chain-reversal loop. This long loop is anchored to the 5'-end of the sequence by an A·T Watson-Crick base pair and a potential G·A noncanonical base pair. These base pairs contribute to the stability of the overall G-quadruplexstructure, as measured by an increase of about 17 kcal/mol in enthalpy or 6 °C in melting temperature. Further, we demonstrate that such a monomorphic structure is formed within longer sequence contexts folding into a pearl-necklace structure.
- G4 notes
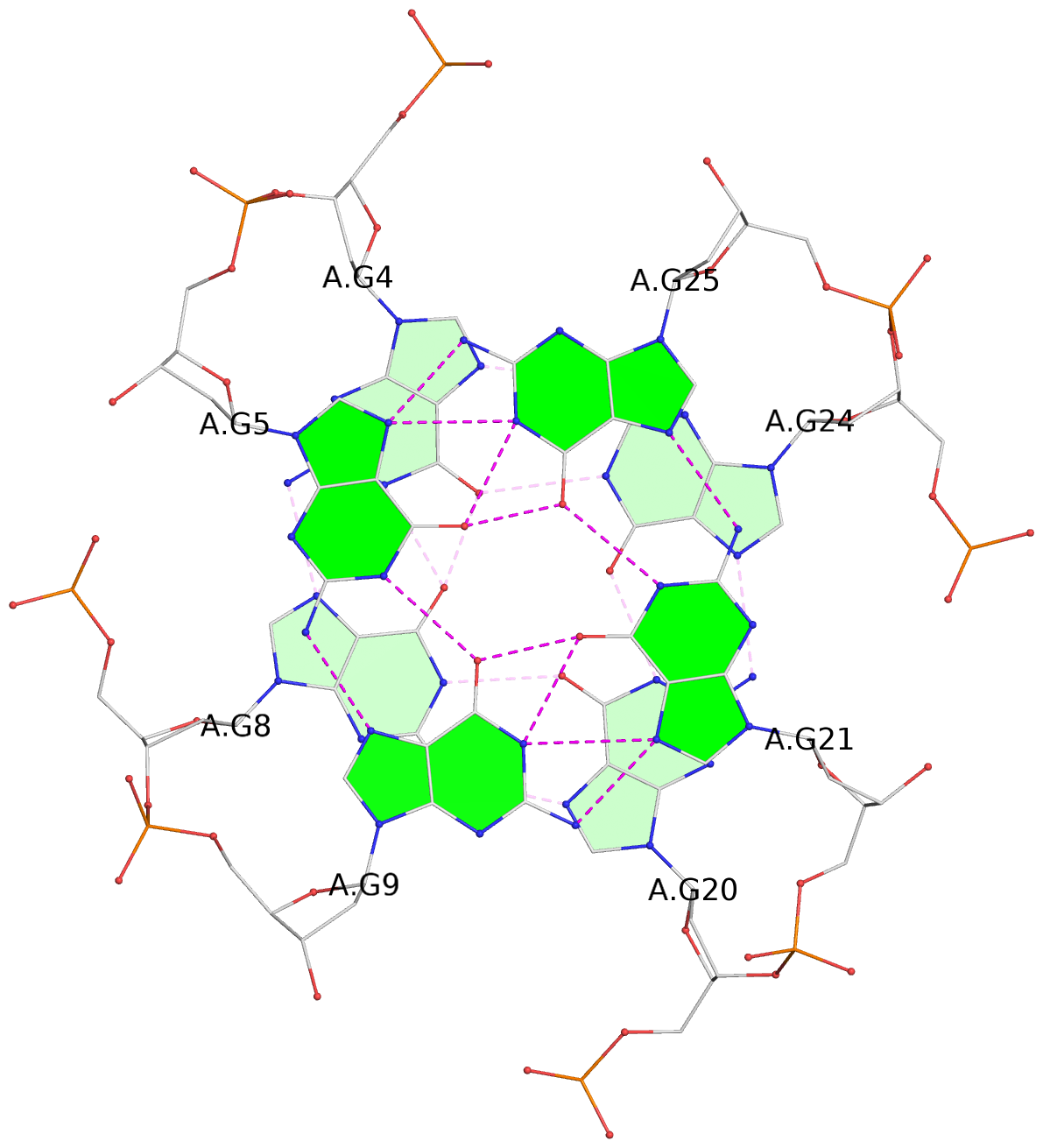
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(-P-P-P), parallel(4+0), UUUU
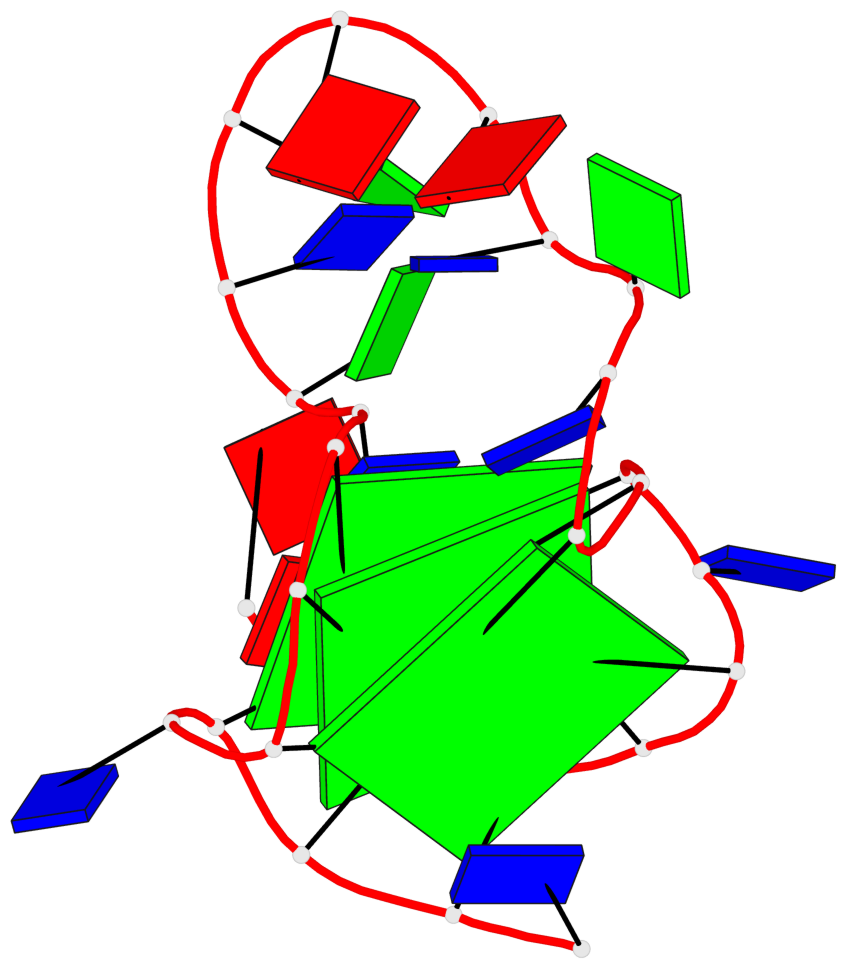
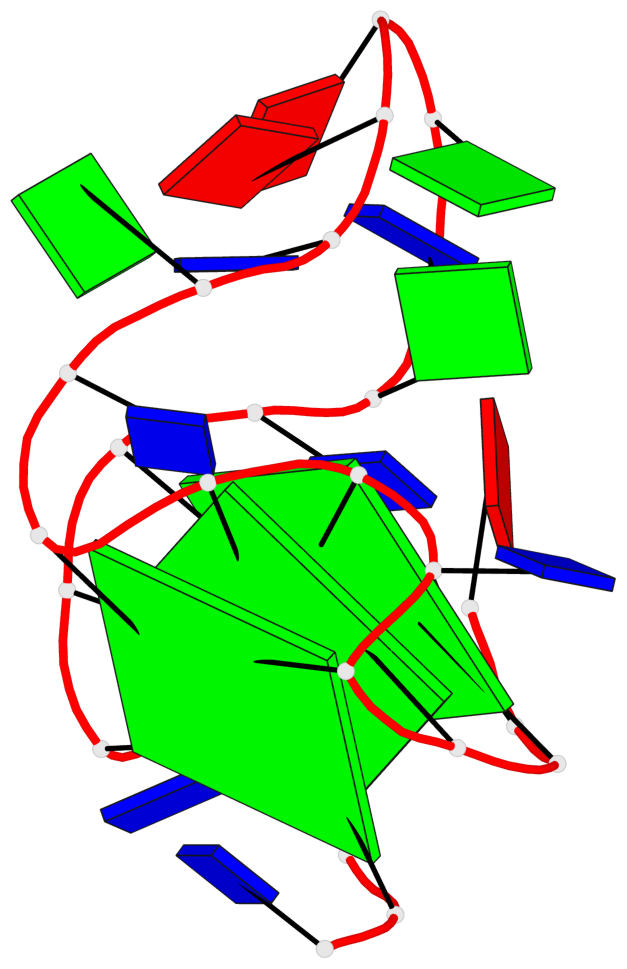
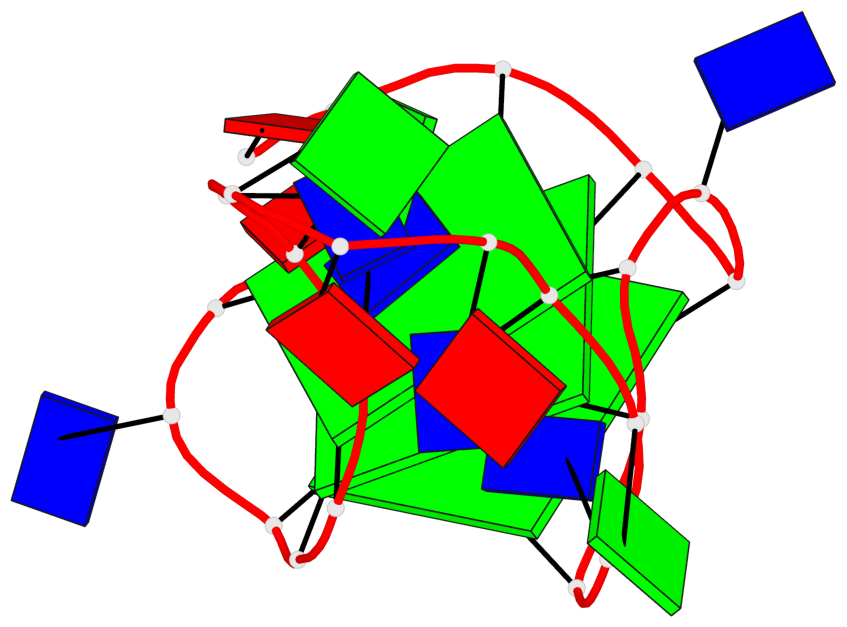
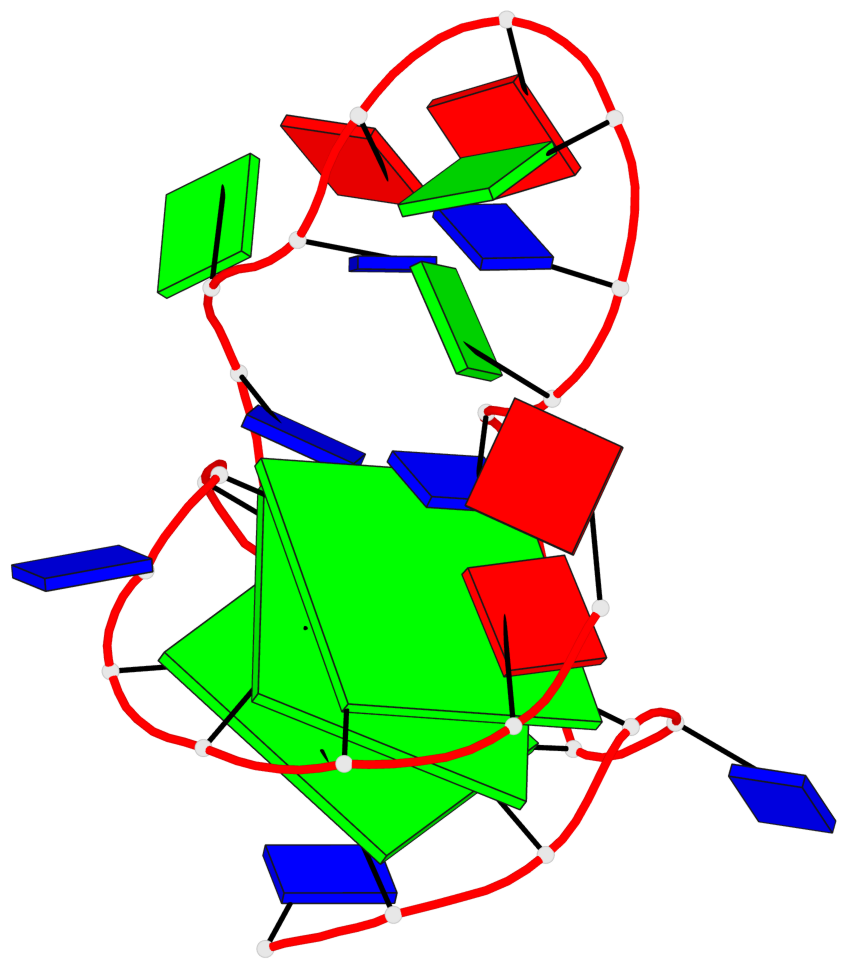
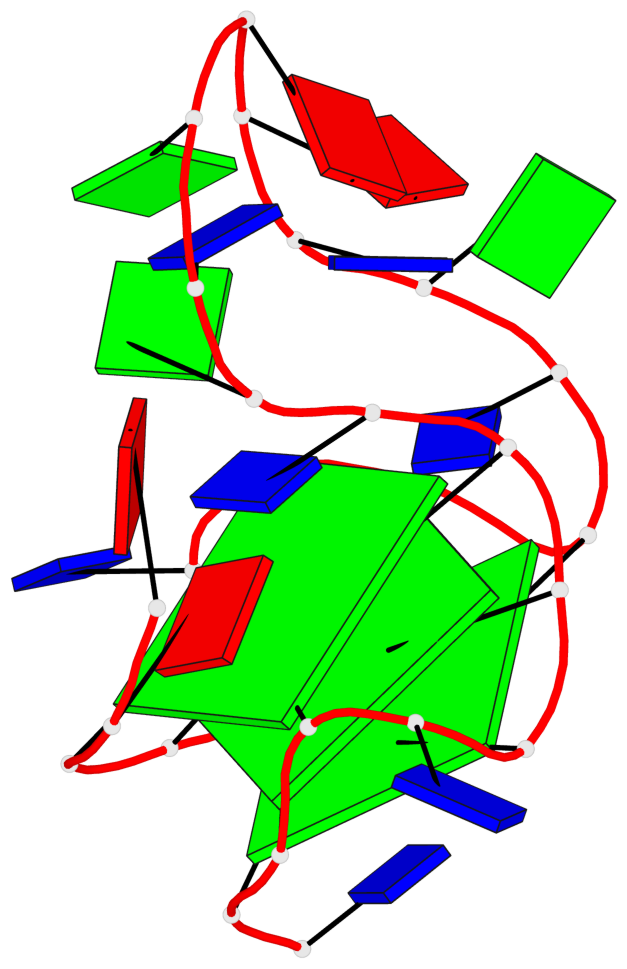
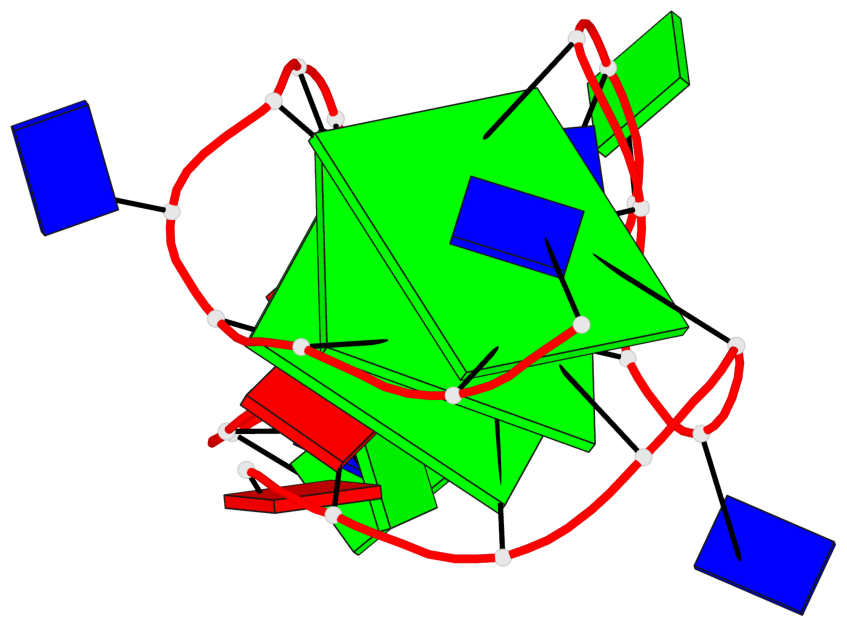
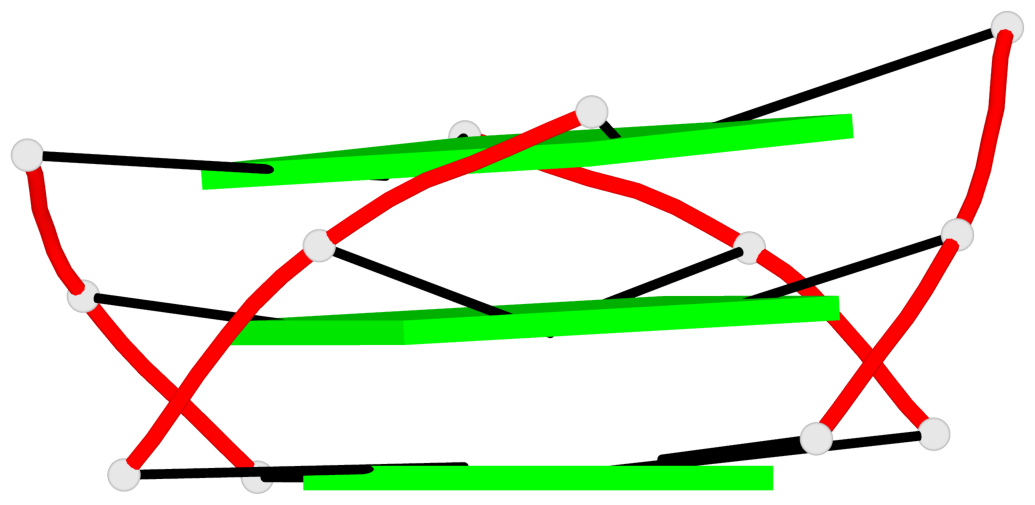
Base-block schematics in six views
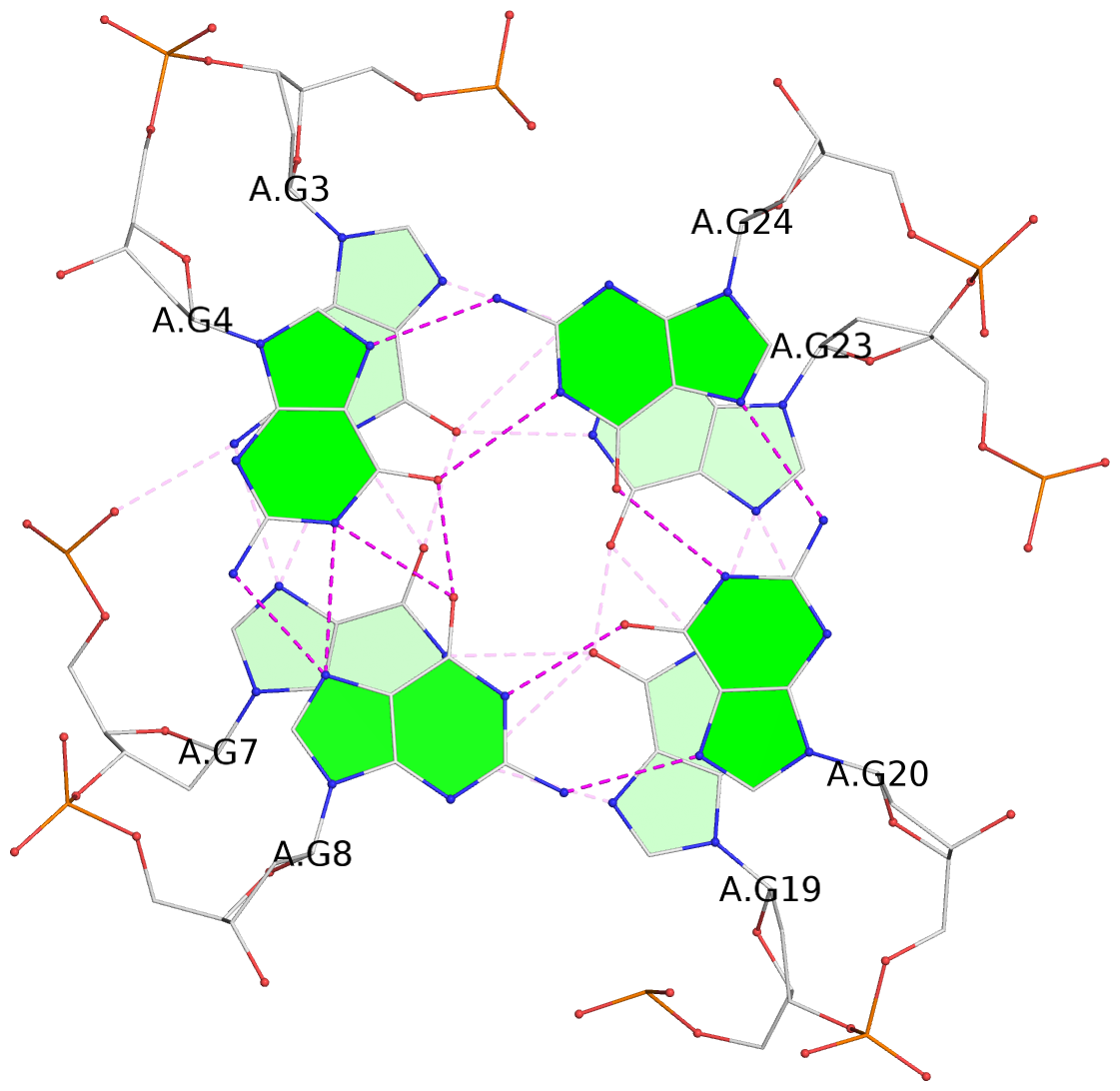
List of 3 G-tetrads
1 glyco-bond=---- sugar=-3.3 groove=---- planarity=0.313 type=other nts=4 GGGG A.DG3,A.DG7,A.DG19,A.DG23 2 glyco-bond=---- sugar=---- groove=---- planarity=0.205 type=other nts=4 GGGG A.DG4,A.DG8,A.DG20,A.DG24 3 glyco-bond=---- sugar=---- groove=---- planarity=0.165 type=other nts=4 GGGG A.DG5,A.DG9,A.DG21,A.DG25
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.