Detailed DSSR results for the G-quadruplex: PDB entry 2lxv
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 2lxv
- Class
- DNA
- Method
- NMR
- Summary
- Dimeric pil-e g-quadruplex DNA from neisseria gonorrhoeae, NMR 11 structures
- Reference
- Kuryavyi V, Cahoon LA, Seifert HS, Patel DJ (2012): "RecA-Binding pilE G4 Sequence Essential for Pilin Antigenic Variation Forms Monomeric and 5' End-Stacked Dimeric Parallel G-Quadruplexes." Structure, 20, 2090-2102. doi: 10.1016/j.str.2012.09.013.
- Abstract
- Neisseria gonorrhoeae is an obligate human pathogen that can escape immune surveillance through antigenic variation of surface structures such as pili. A G-quadruplex-forming (G4) sequence (5'-G(3)TG(3)TTG(3)TG(3)) located upstream of the N. gonorrhoeae pilin expression locus (pilE) is necessary for initiation of pilin antigenic variation, a recombination-based, high-frequency, diversity-generation system. We have determined NMR-based structures of the all parallel-stranded monomeric and 5' end-stacked dimeric pilE G-quadruplexes in monovalent cation-containing solutions. We demonstrate that the three-layered all parallel-stranded monomeric pilE G-quadruplex containing single-residue double-chain reversal loops, which can be modeled without steric clashes into the 3 nt DNA-binding site of RecA, binds and promotes E. coli RecA-mediated strand exchange in vitro. We discuss how interactions between RecA and monomeric pilE G-quadruplex could facilitate the specialized recombination reactions leading to pilin diversification.
- G4 notes
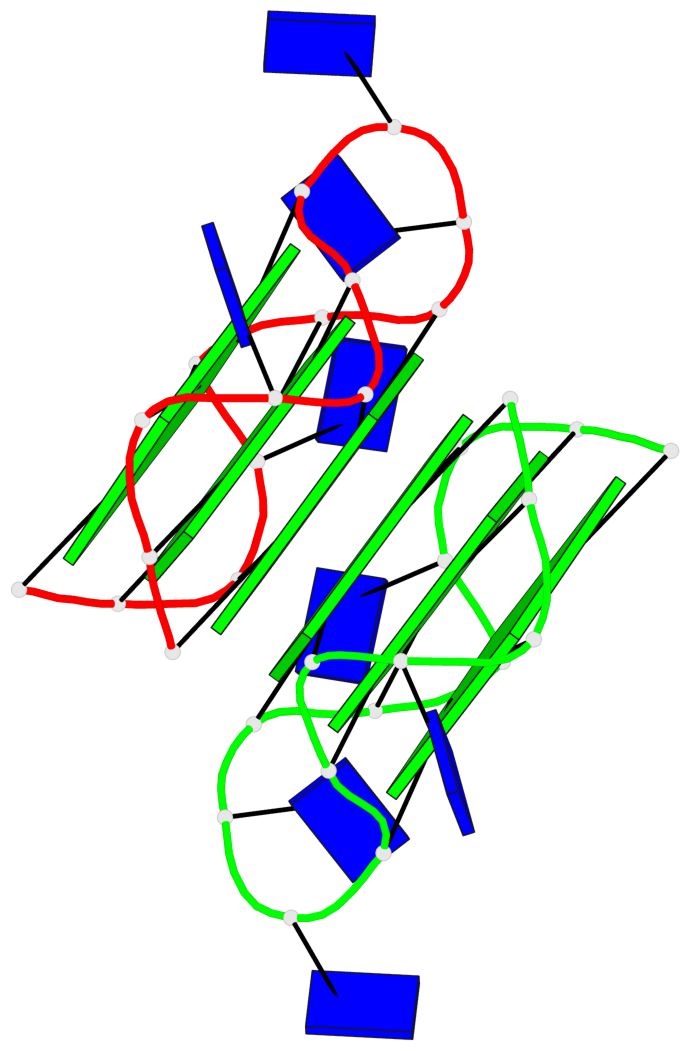
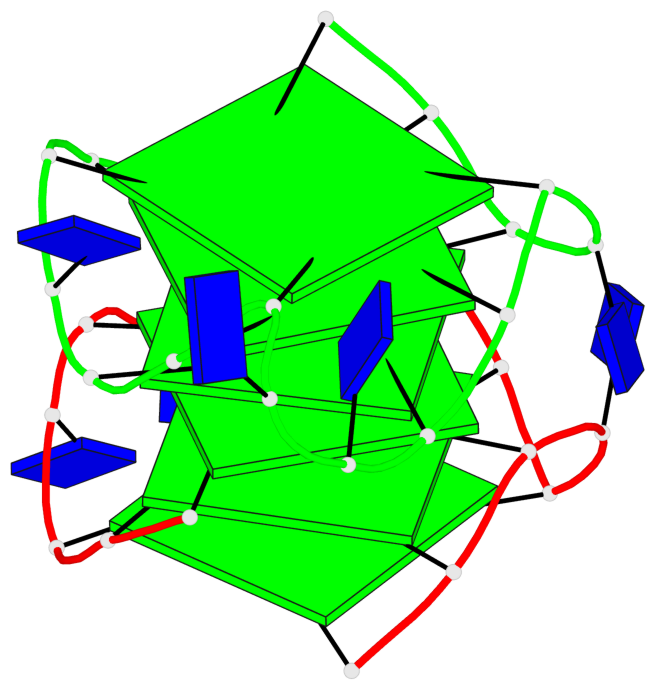
- 6 G-tetrads, 1 G4 helix, 2 G4 stems, 1 G4 coaxial stack, 3(-P-P-P), parallel(4+0), UUUU, coaxial interfaces: 5'/5'
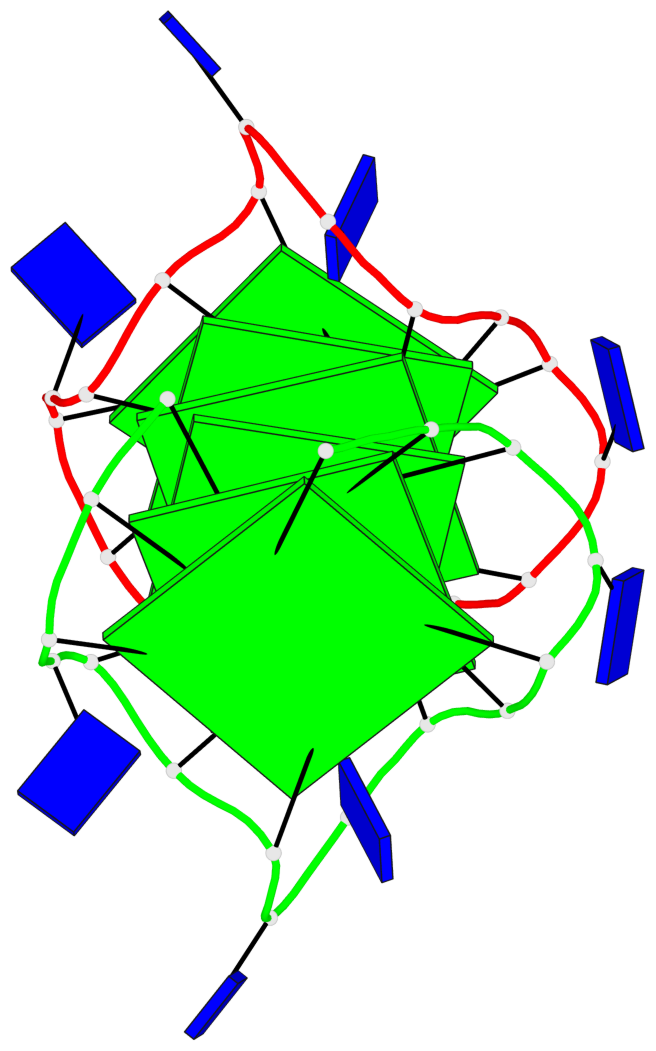
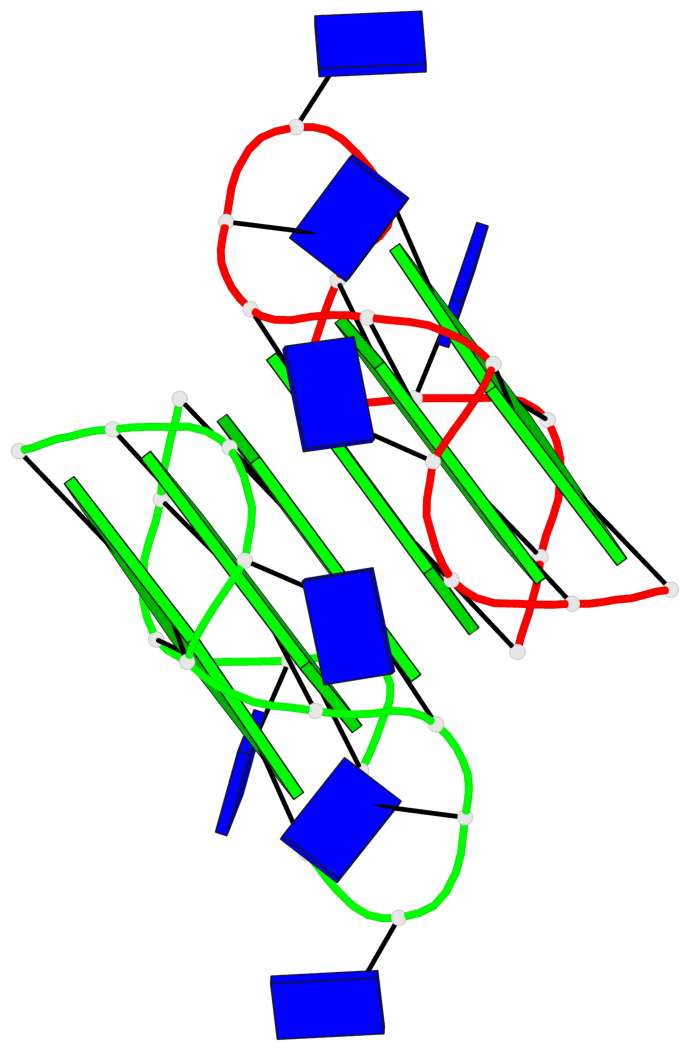
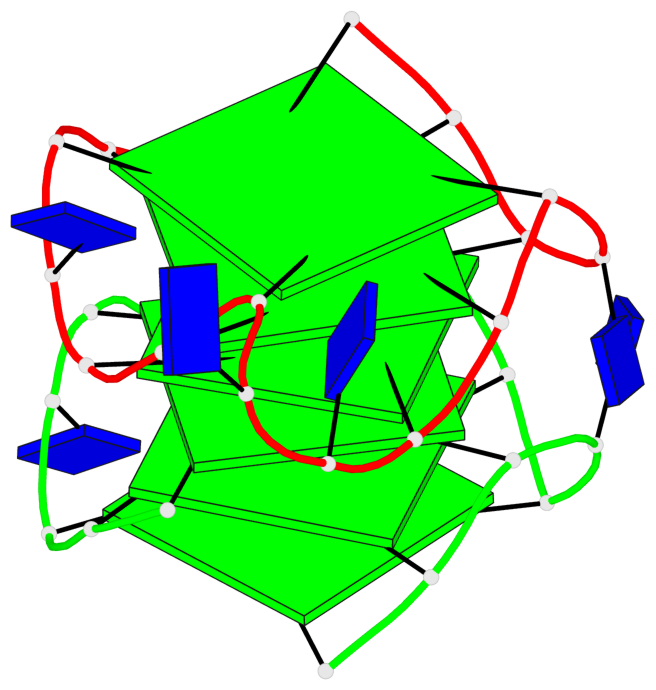
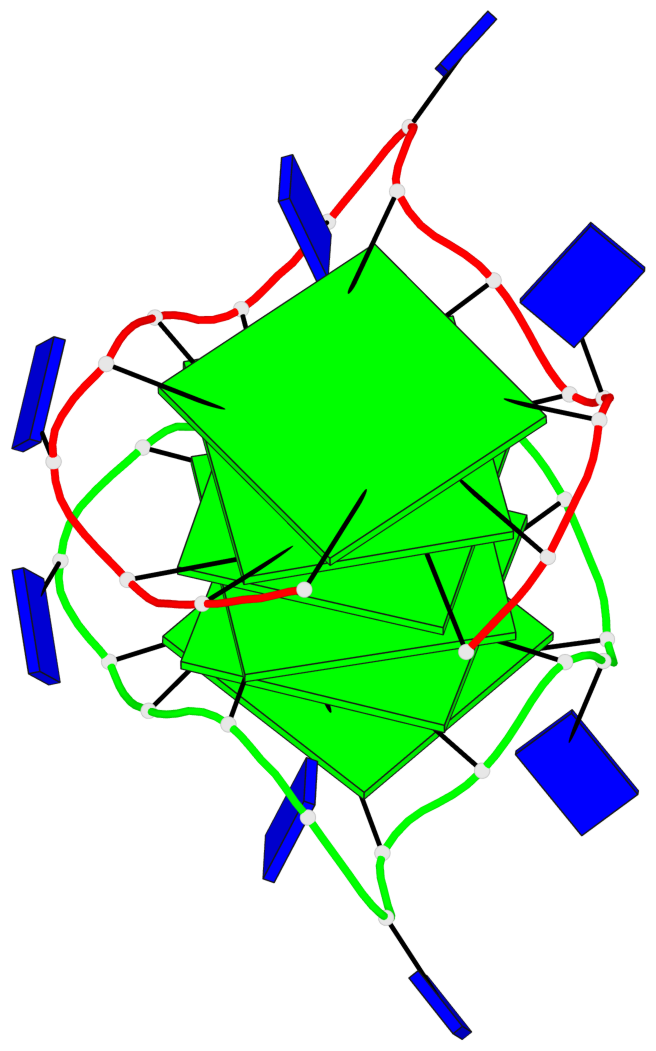
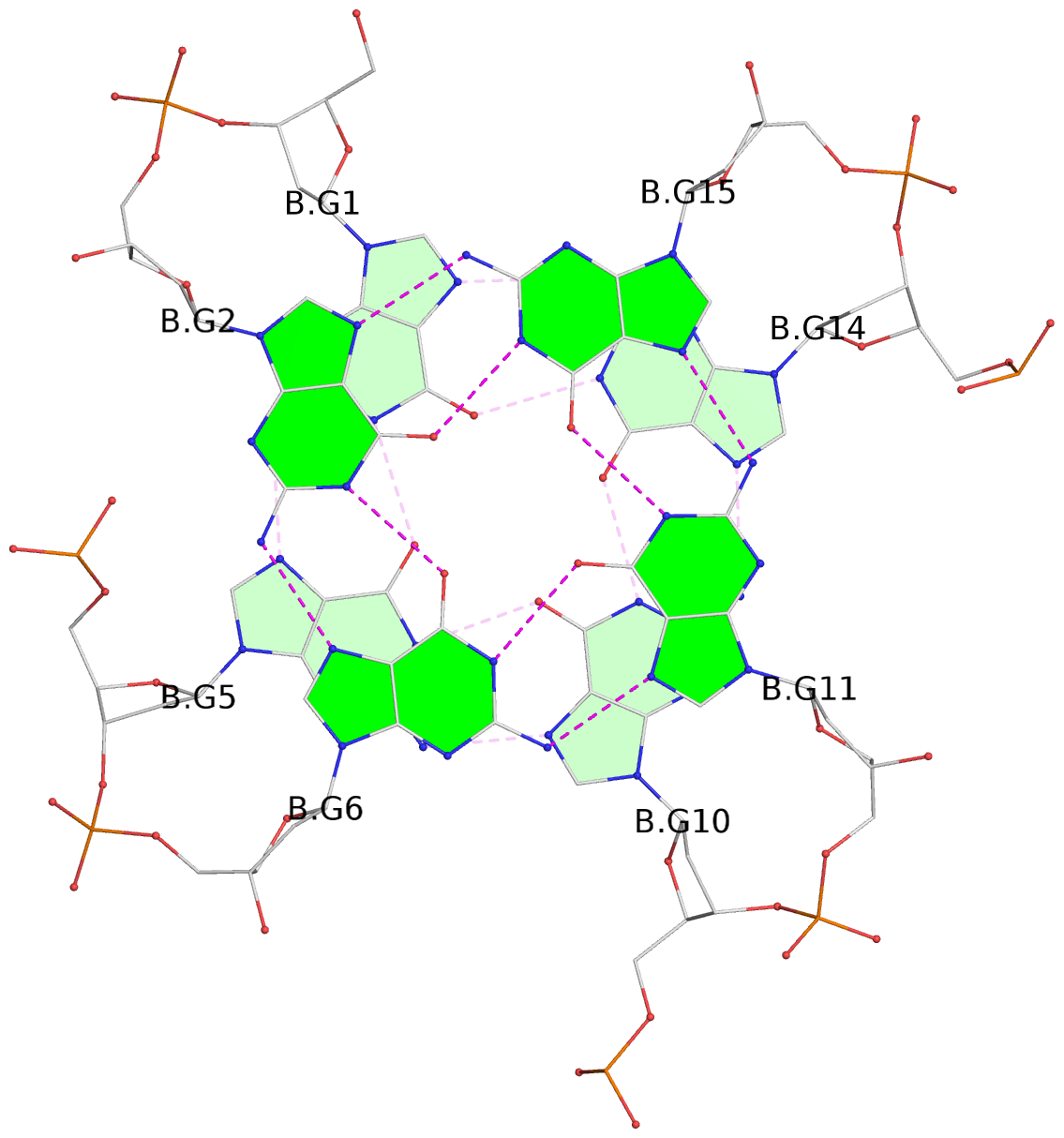
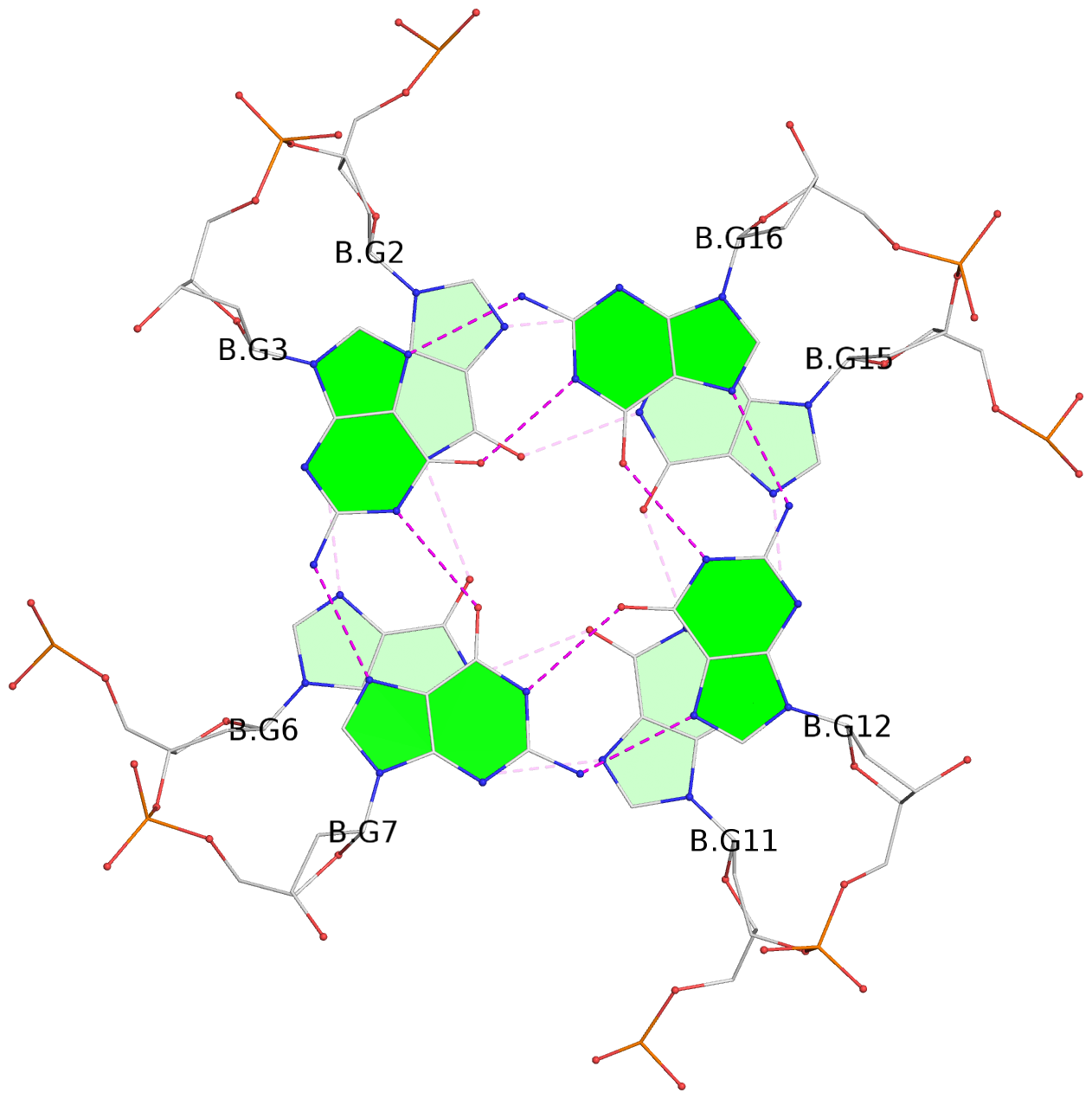
Base-block schematics in six views
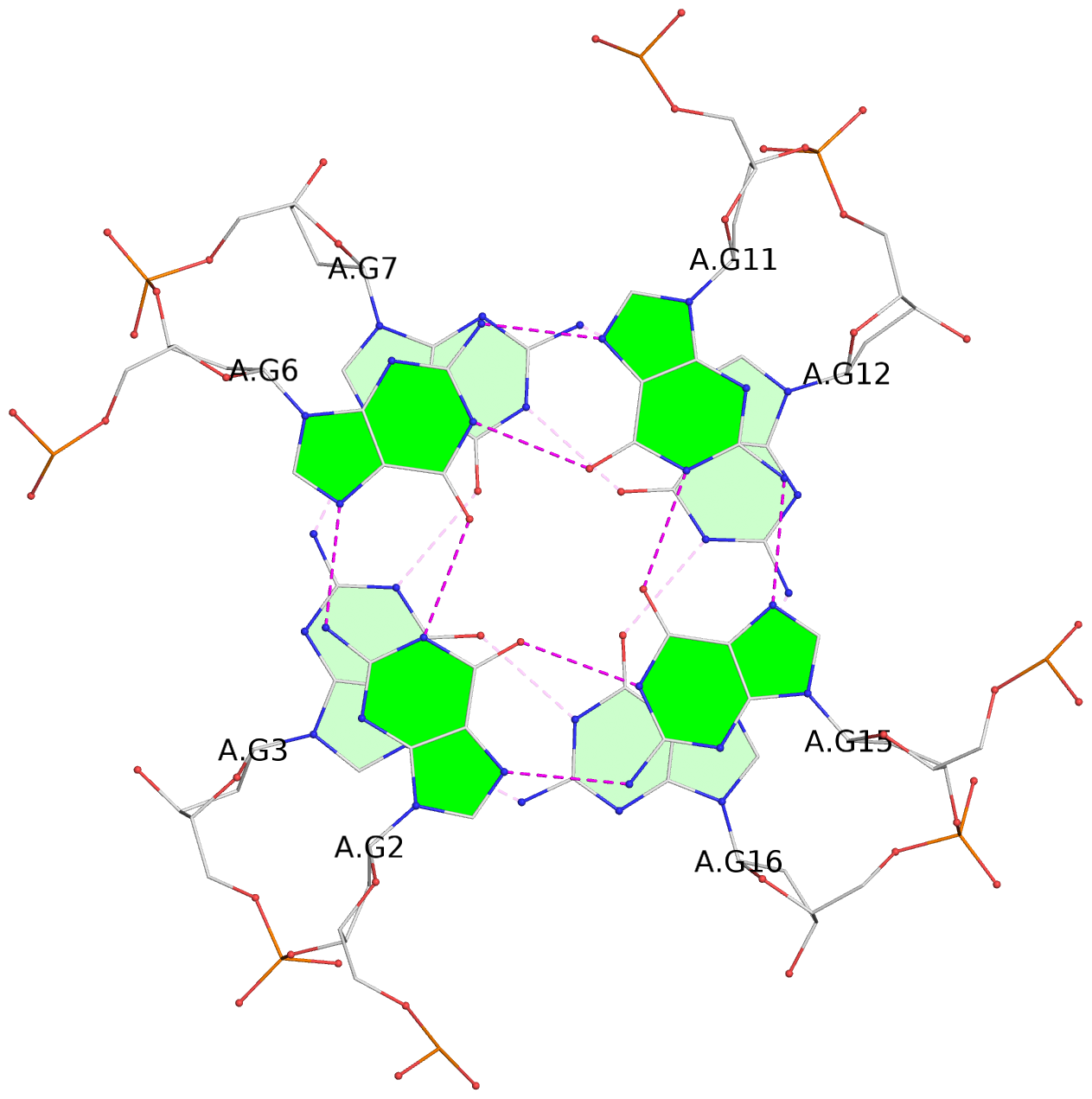
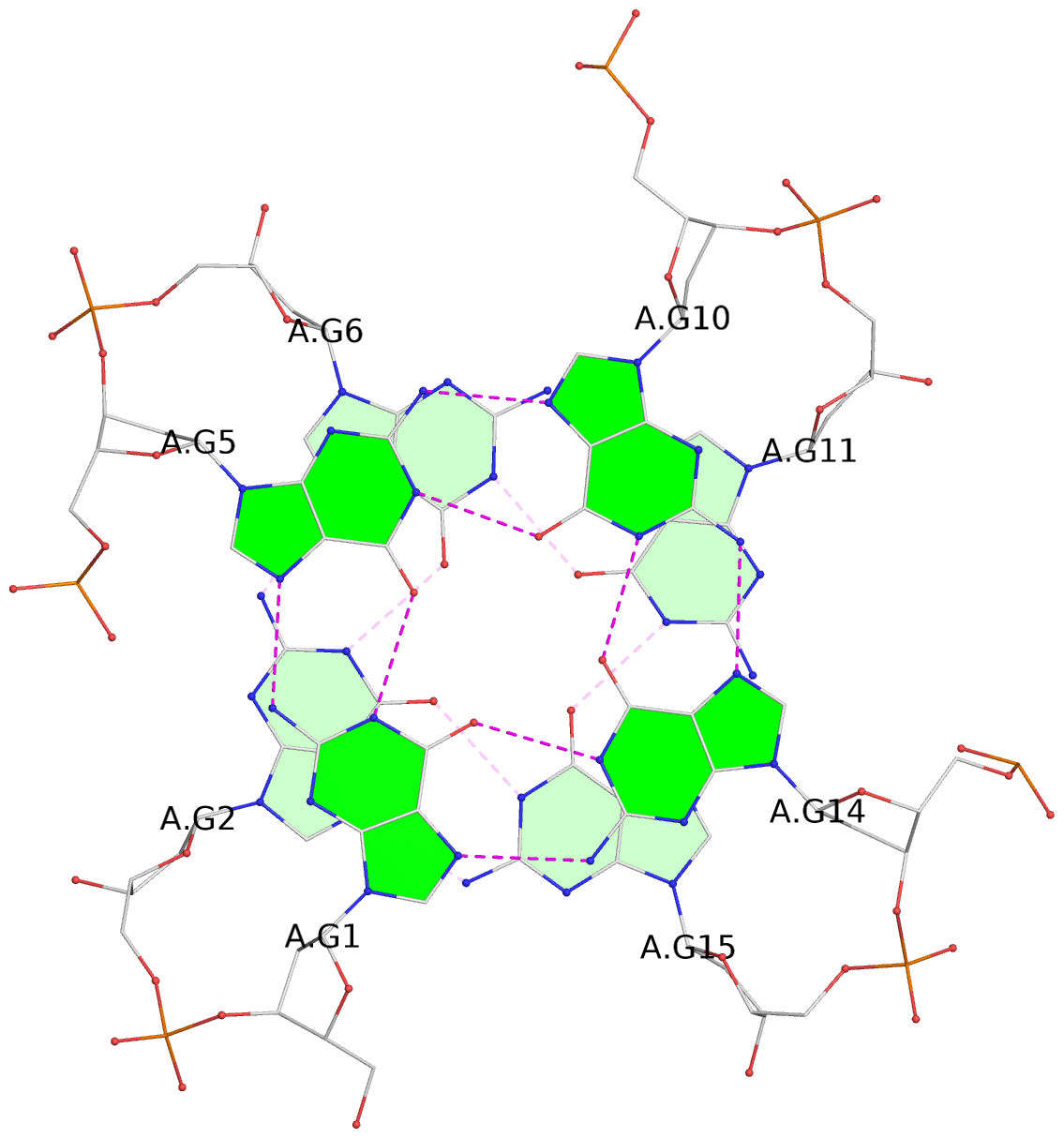
List of 6 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.129 type=planar nts=4 GGGG A.DG1,A.DG5,A.DG10,A.DG14 2 glyco-bond=---- sugar=.... groove=---- planarity=0.064 type=planar nts=4 GGGG A.DG2,A.DG6,A.DG11,A.DG15 3 glyco-bond=---- sugar=---- groove=---- planarity=0.165 type=other nts=4 GGGG A.DG3,A.DG7,A.DG12,A.DG16 4 glyco-bond=---- sugar=---- groove=---- planarity=0.130 type=planar nts=4 GGGG B.DG1,B.DG5,B.DG10,B.DG14 5 glyco-bond=---- sugar=.... groove=---- planarity=0.064 type=planar nts=4 GGGG B.DG2,B.DG6,B.DG11,B.DG15 6 glyco-bond=---- sugar=---- groove=---- planarity=0.165 type=other nts=4 GGGG B.DG3,B.DG7,B.DG12,B.DG16
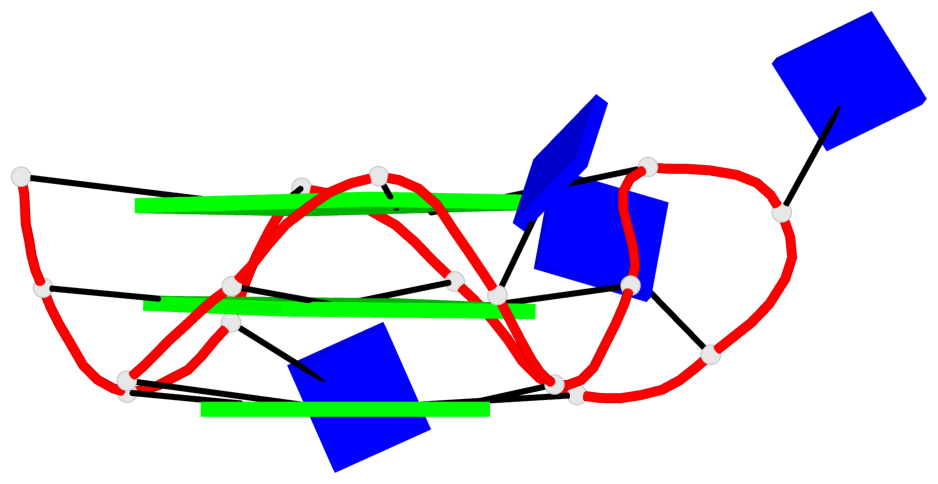
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 6 G-tetrad layers, inter-molecular, with 2 stems
List of 2 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 3 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 3(-P-P-P), parallel(4+0)
Stem#2, 3 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 3(-P-P-P), parallel(4+0)
List of 1 G4 coaxial stack
1 G4 helix#1 contains 2 G4 stems: [#1,#2] [5'/5']