Detailed DSSR results for the G-quadruplex: PDB entry 2m18
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 2m18
- Class
- RNA
- Method
- NMR
- Summary
- Structure of stacked g-quadruplex formed by human terra sequence in potassium solution
- Reference
- Martadinata H, Phan AT (2013): "Structure of Human Telomeric RNA (TERRA): Stacking of Two G-Quadruplex Blocks in K(+) Solution." Biochemistry, 52, 2176-2183. doi: 10.1021/bi301606u.
- Abstract
- Telomeric repeat-containing RNAs (TERRA) are transcription products of the telomeres. Human TERRA sequences containing UUAGGG repeats can form parallel-stranded G-quadruplexes. The stacking interaction of such structures was shown to be important for ligand targeting and higher-order arrangement of G-quadruplexes in long TERRA sequences. Here we report on the first high-resolution structure of a stacked G-quadruplex formed by the 10-nucleotide human TERRA sequence r(GGGUUAGGGU) in potassium solution. This structure comprises two dimeric three-layer parallel-stranded G-quadruplex blocks, which stack on each other at their 5'-ends. The adenine in each UUA loop is nearly coplanar with the 5'-end G-tetrad forming an A·(G·G·G·G)·A hexad, thereby increasing the stacking contacts between the two blocks. Interestingly, this stacking and loop conformation is different from all structures previously reported for the free human TERRA but resembles the structure previously determined for a complex between a human TERRA sequence and an acridine ligand. This stacking conformation is a potential target for drugs that recognize or induce the stacking interface.
- G4 notes
- 6 G-tetrads, 1 G4 helix, 2 G4 stems, 1 G4 coaxial stack, parallel(4+0), UUUU, coaxial interfaces: 5'/5'
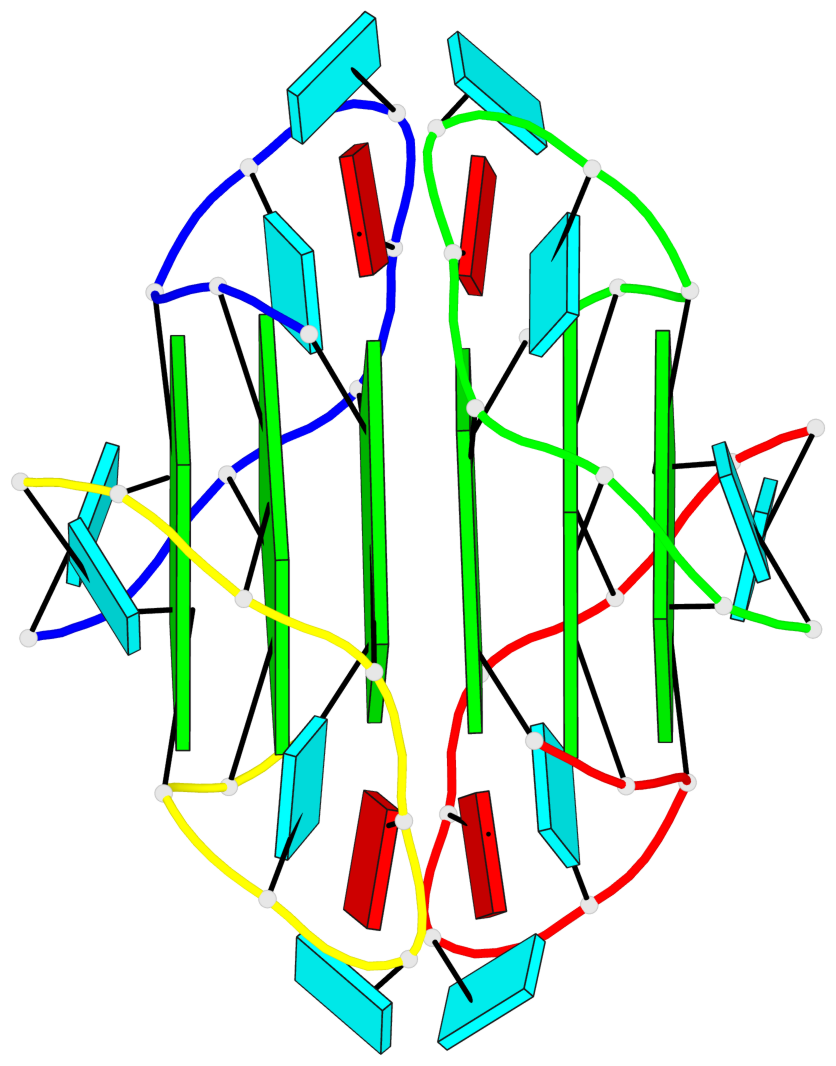
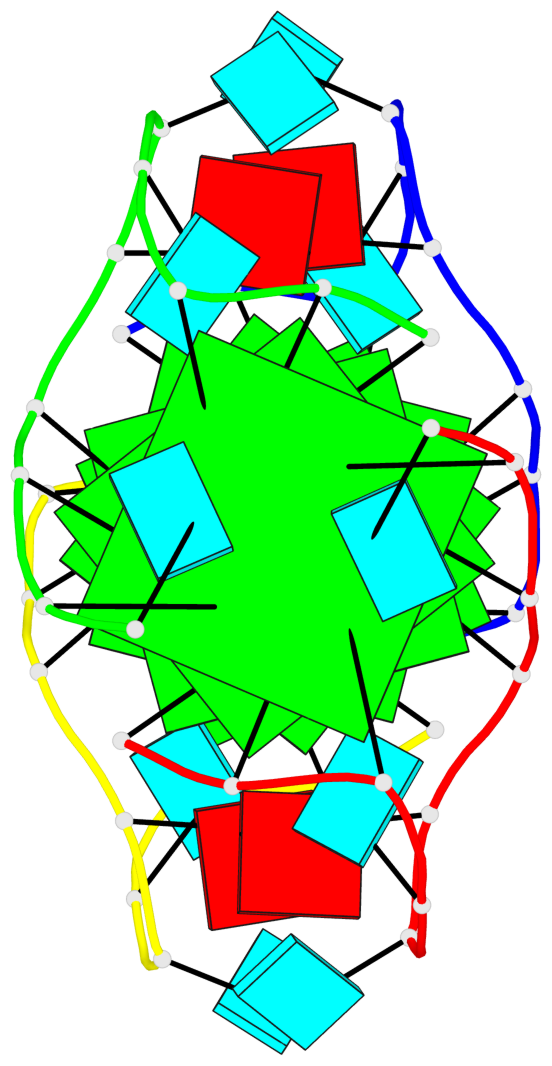
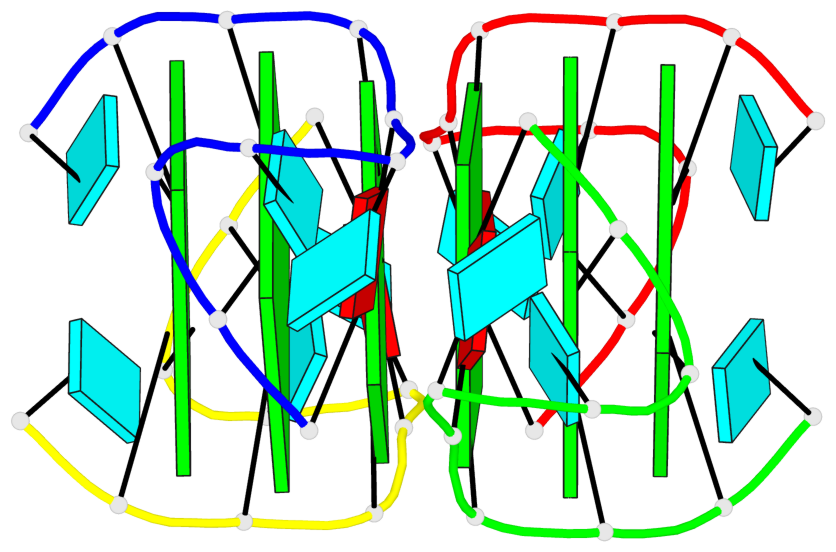
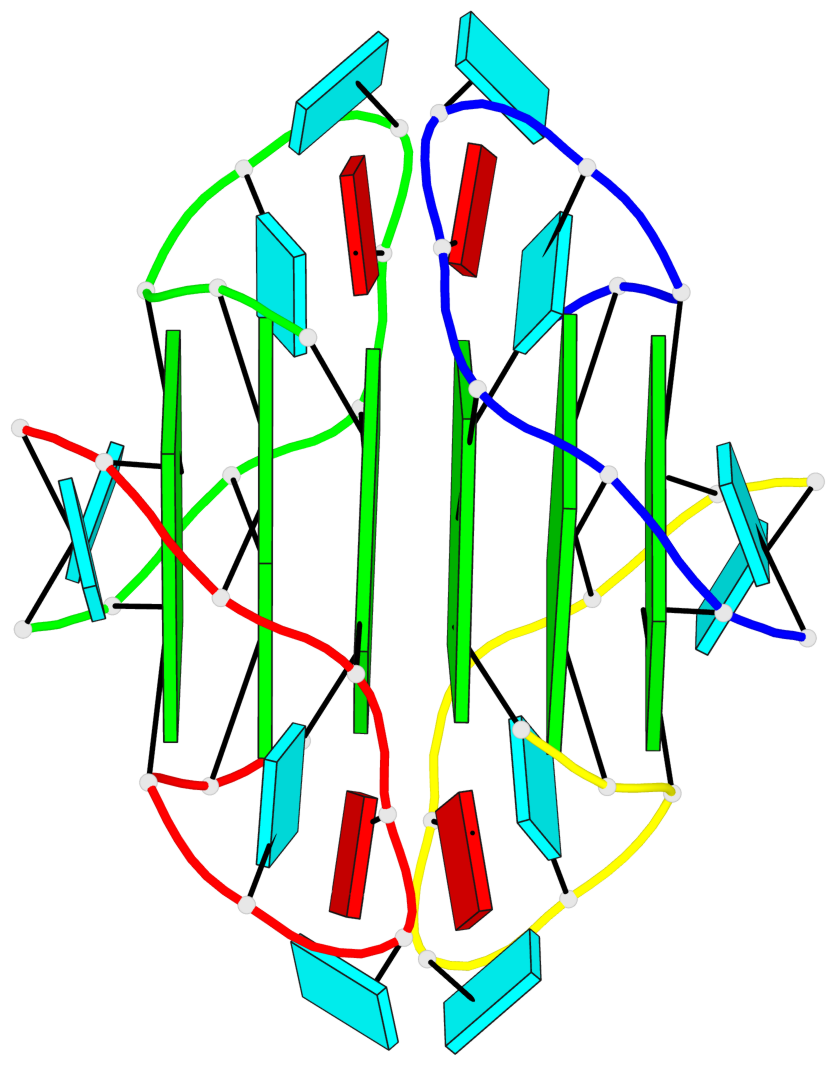
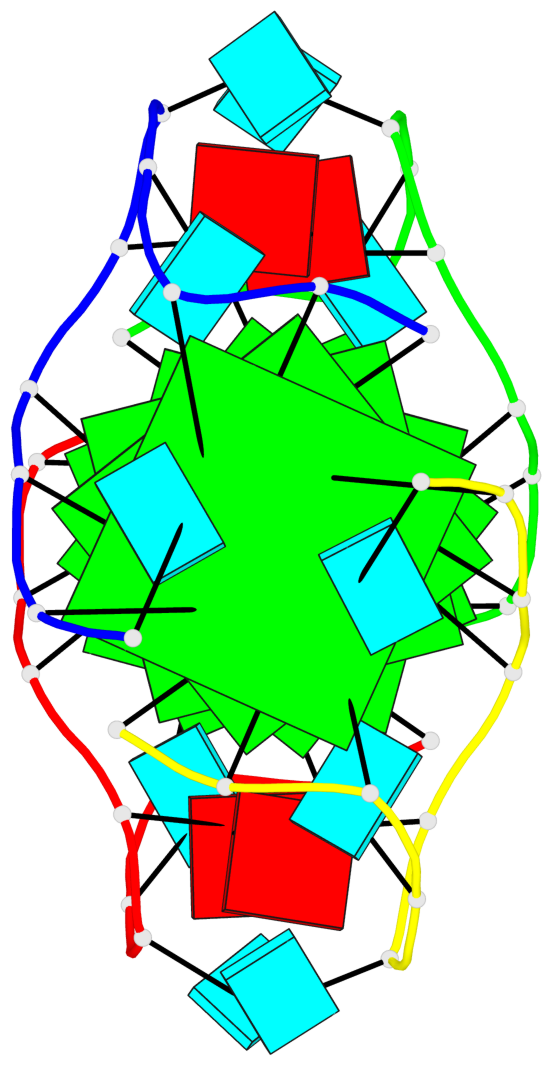
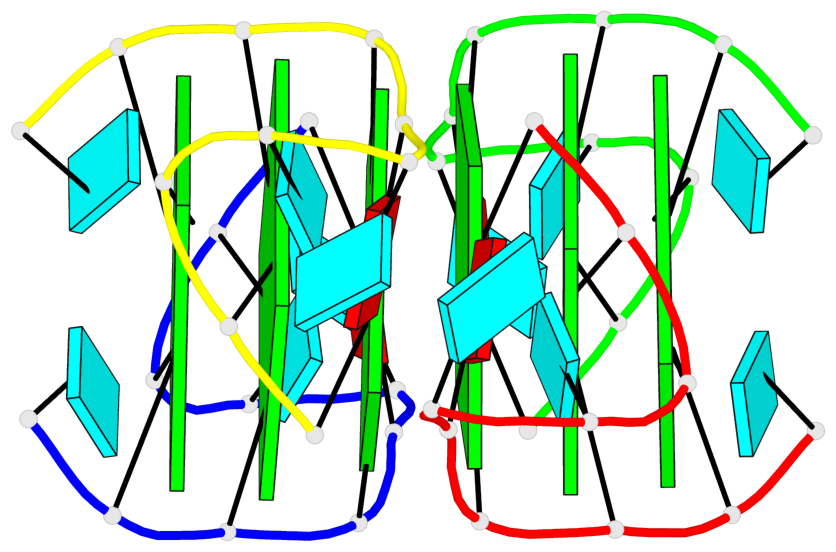
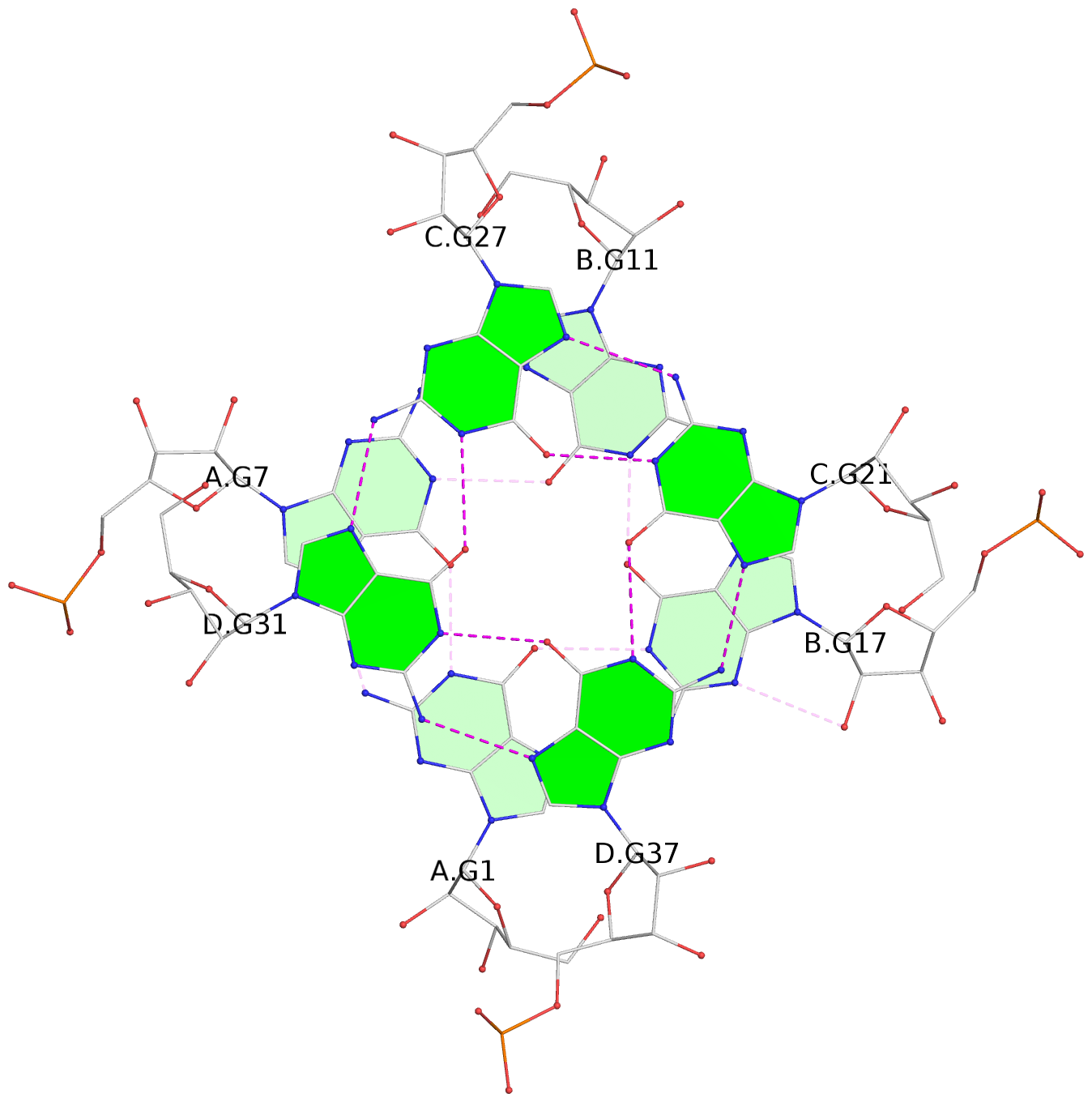
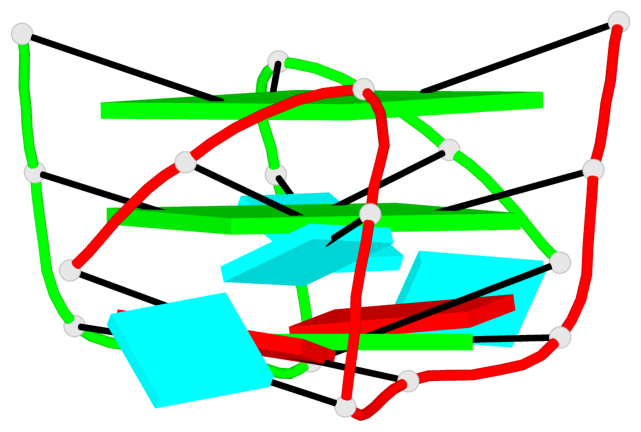
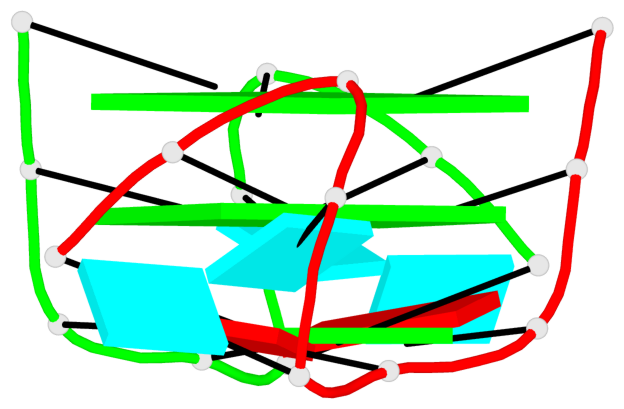
Base-block schematics in six views
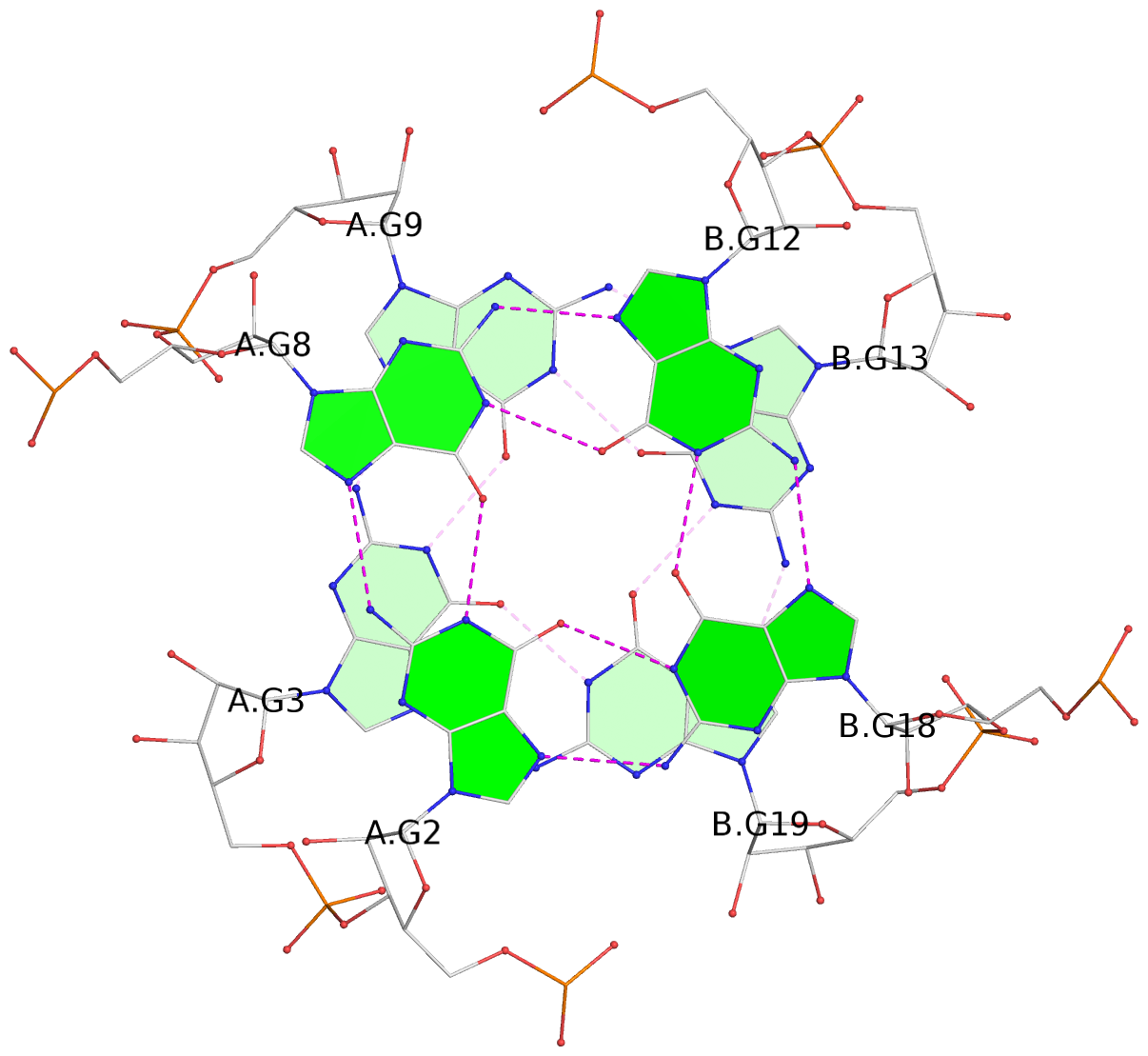
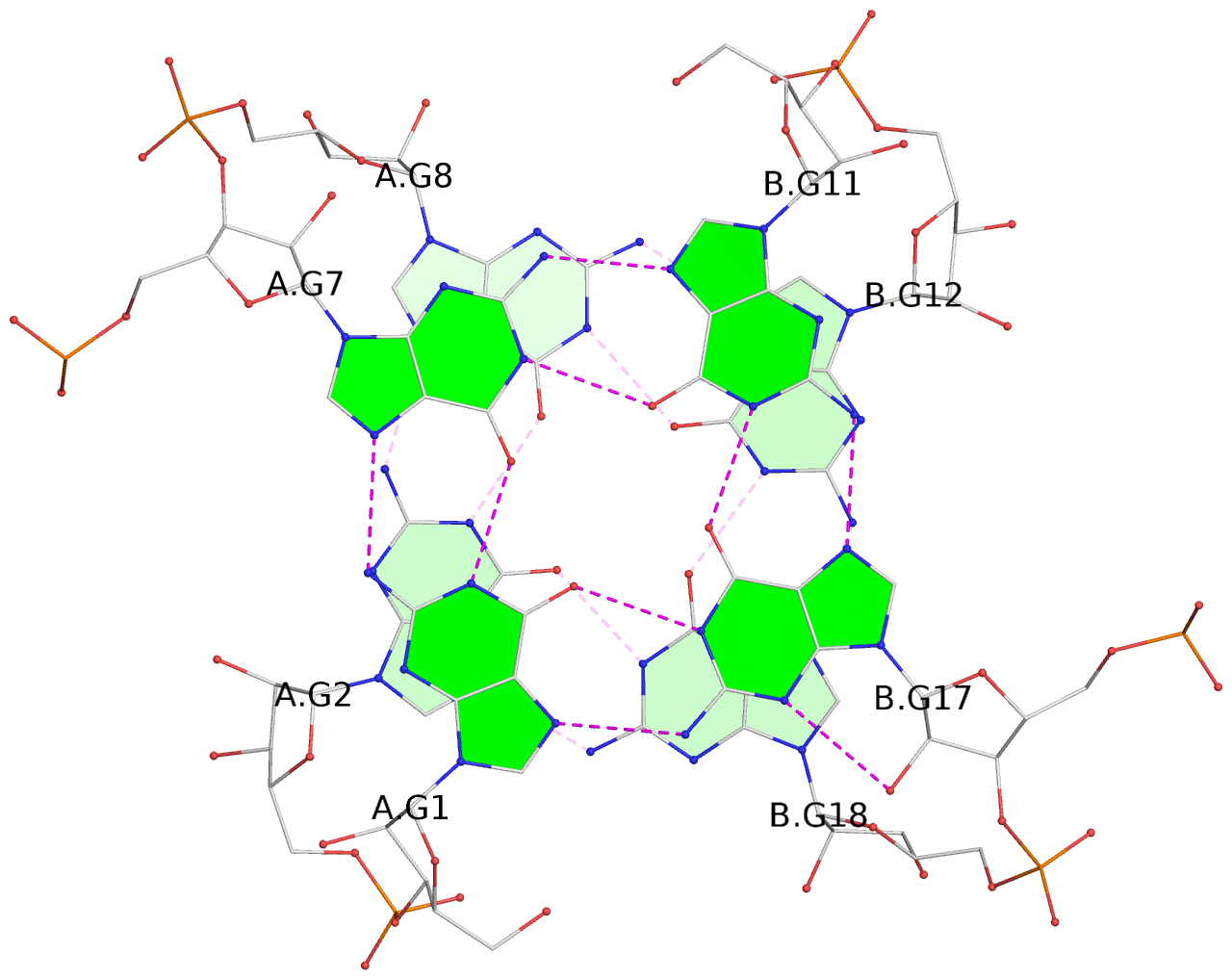
List of 6 G-tetrads
1 glyco-bond=---- sugar=3.3. groove=---- planarity=0.494 type=other nts=4 GGGG A.G1,A.G7,B.G11,B.G17 2 glyco-bond=---- sugar=3333 groove=---- planarity=0.419 type=saddle nts=4 GGGG A.G2,A.G8,B.G12,B.G18 3 glyco-bond=---- sugar=3333 groove=---- planarity=0.432 type=other nts=4 GGGG A.G3,A.G9,B.G13,B.G19 4 glyco-bond=---- sugar=3.3- groove=---- planarity=0.494 type=other nts=4 GGGG C.G21,C.G27,D.G31,D.G37 5 glyco-bond=---- sugar=3333 groove=---- planarity=0.487 type=other nts=4 GGGG C.G22,C.G28,D.G32,D.G38 6 glyco-bond=---- sugar=3333 groove=---- planarity=0.498 type=other nts=4 GGGG C.G23,C.G29,D.G33,D.G39
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 6 G-tetrad layers, inter-molecular, with 2 stems
List of 2 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 3 G-tetrad layers, 2 loops, inter-molecular, UUUU, parallel, parallel(4+0)
Stem#2, 3 G-tetrad layers, 2 loops, inter-molecular, UUUU, parallel, parallel(4+0)
List of 1 G4 coaxial stack
1 G4 helix#1 contains 2 G4 stems: [#1,#2] [5'/5']