Detailed DSSR results for the G-quadruplex: PDB entry 2m1g
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 2m1g
- Class
- DNA
- Method
- NMR
- Summary
- Parallel human telomeric quadruplex containing 2'f-ana substitutions
- Reference
- Martin-Pintado N, Yahyaee-Anzahaee M, Deleavey GF, Portella G, Orozco M, Damha MJ, Gonzalez C (2013): "Dramatic effect of furanose c2' substitution on structure and stability: directing the folding of the human telomeric quadruplex with a single fluorine atom." J.Am.Chem.Soc., 135, 5344-5347. doi: 10.1021/ja401954t.
- Abstract
- Human telomeric DNA quadruplexes can adopt different conformations in solution. We have found that arabinose, 2'F-arabinose, and ribose substitutions stabilize the propeller parallel G-quadruplex form over competing conformers, allowing NMR structural determination of this particularly significant nucleic acid structure. 2'F-arabinose substitution provides the greatest stabilization as a result of electrostatic (F-CH---O4') and pseudo-hydrogen-bond (F---H8) stabilizing interactions. In contrast, 2'F-rG substitution provokes a dramatic destabilization of the quadruplex structure due to unfavorable electrostatic repulsion between the phosphate and the 2'-F.
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, parallel(4+0), UUUU
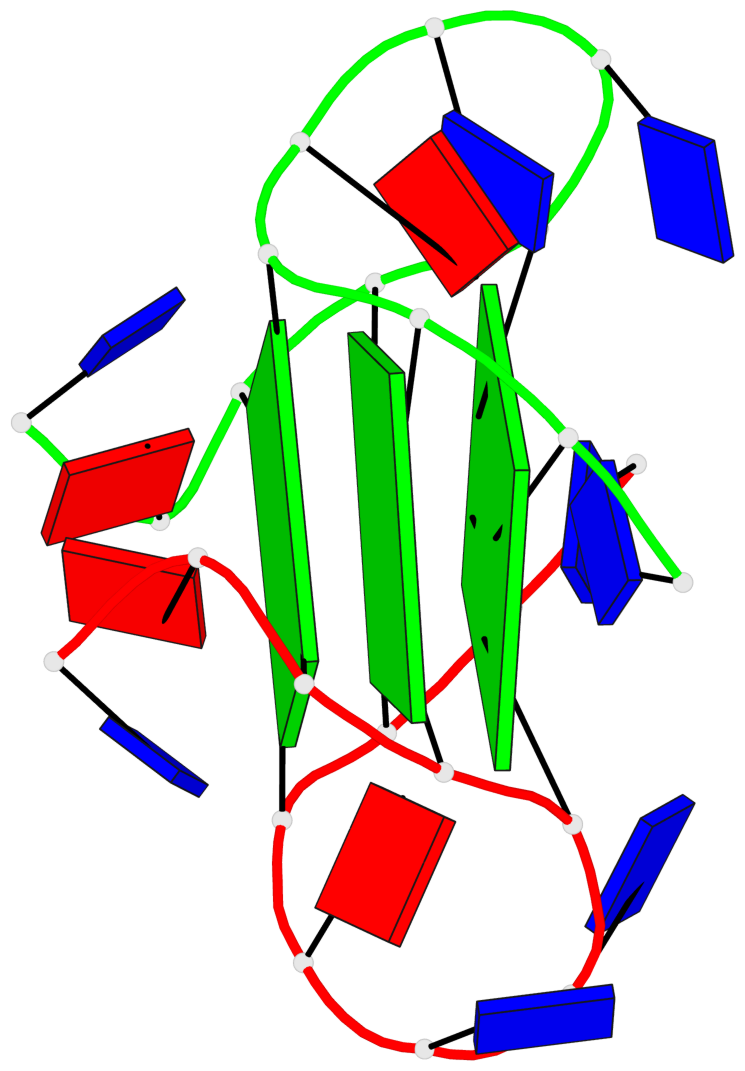
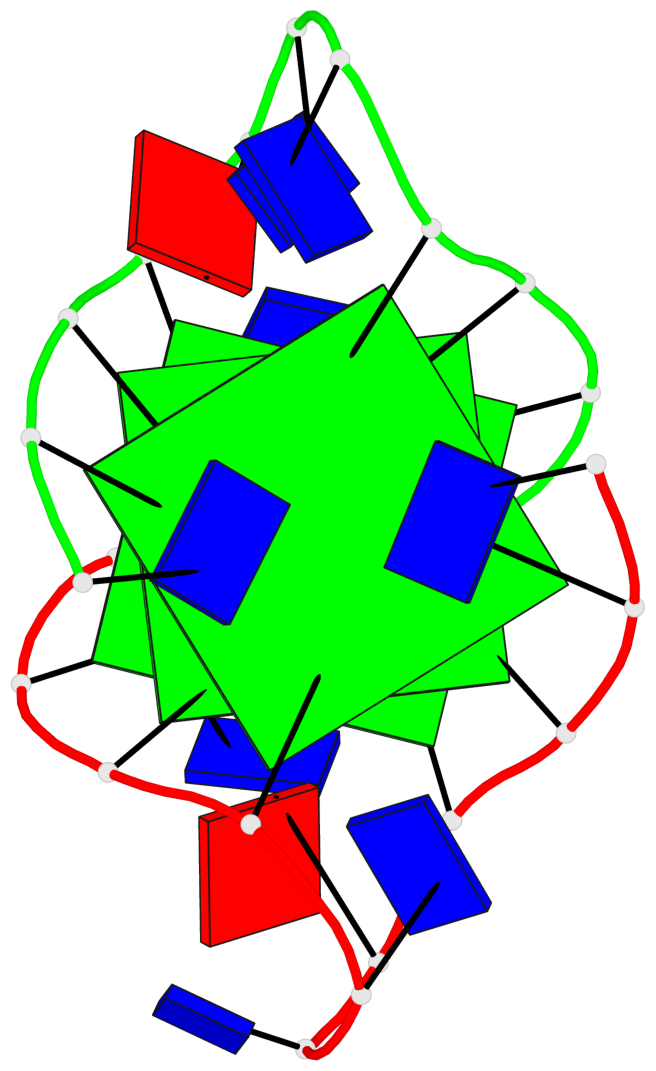
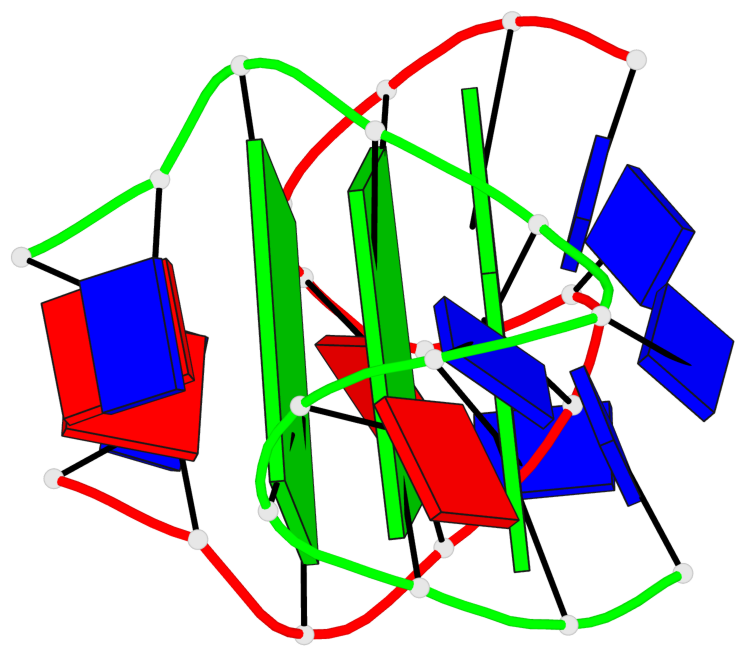
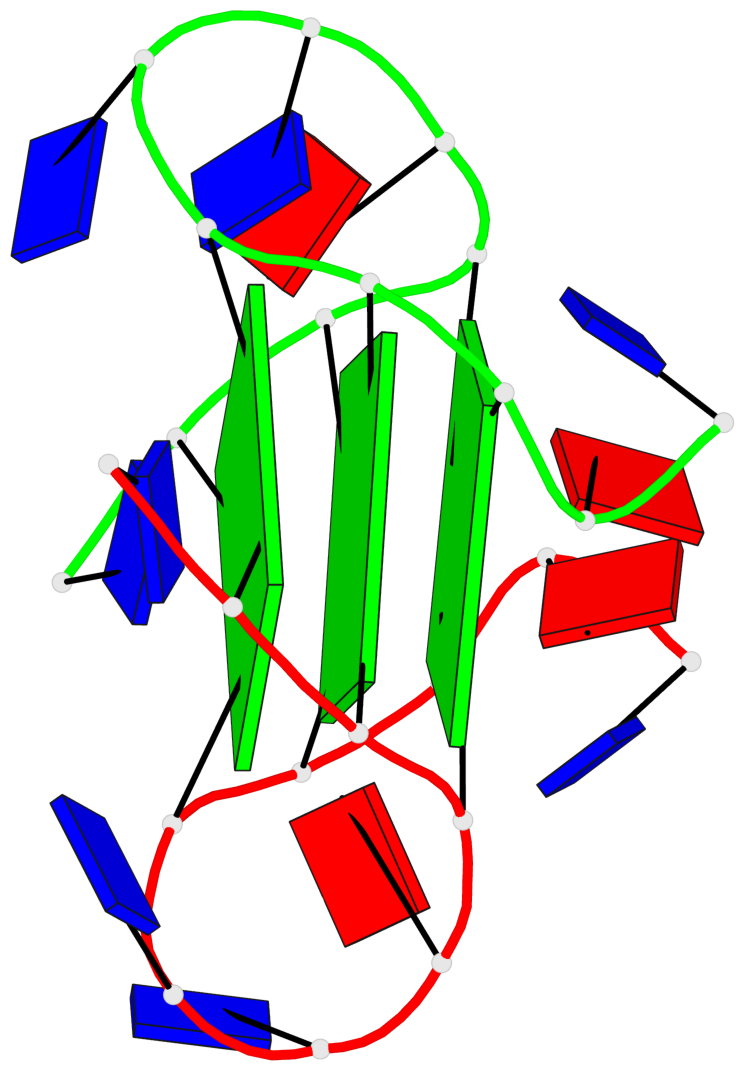
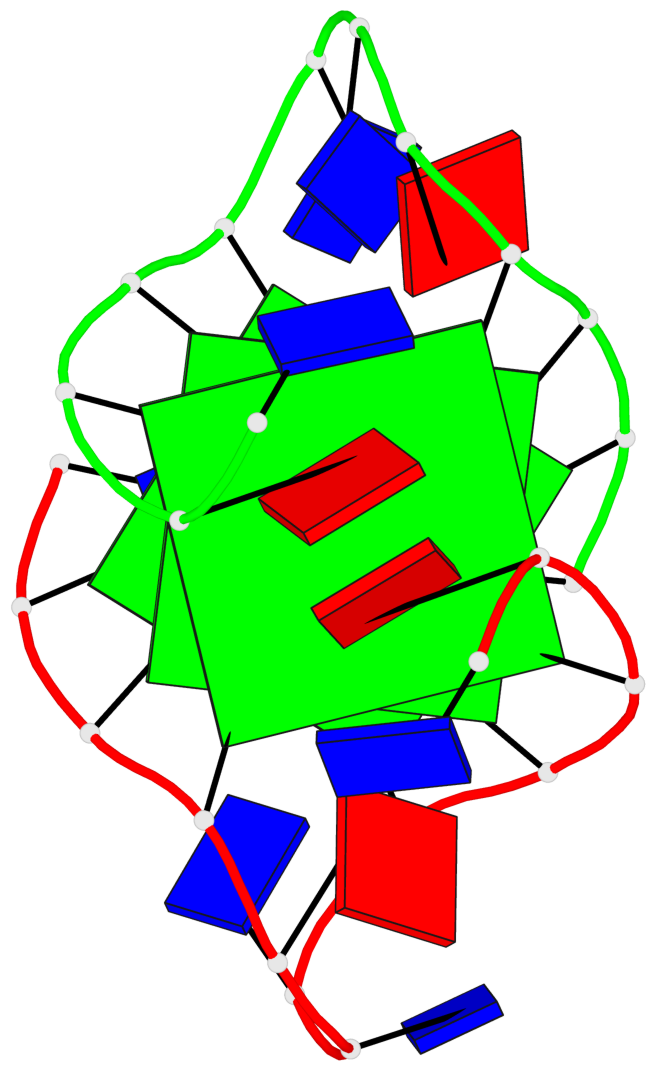
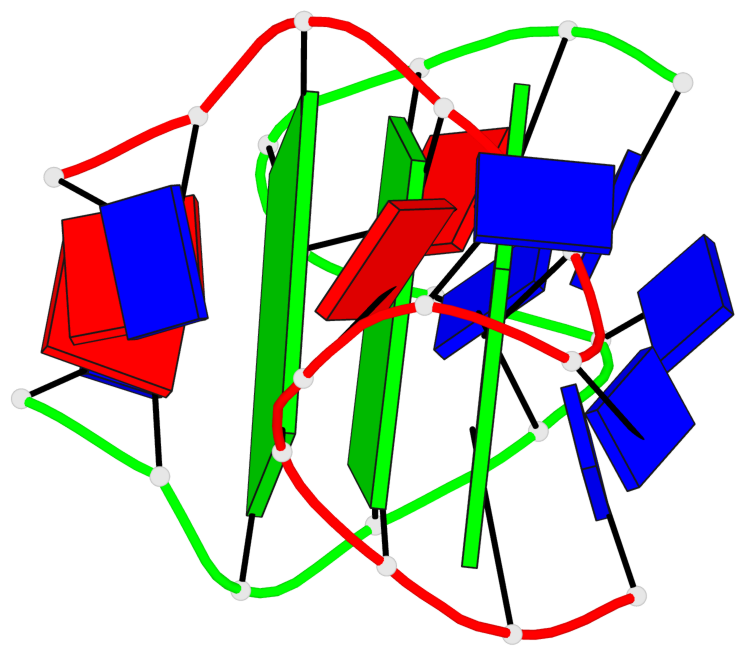
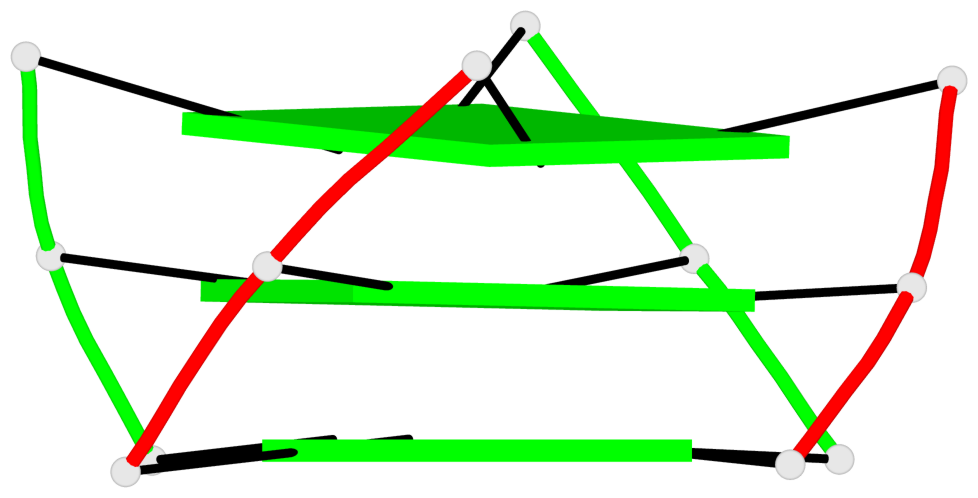
Base-block schematics in six views
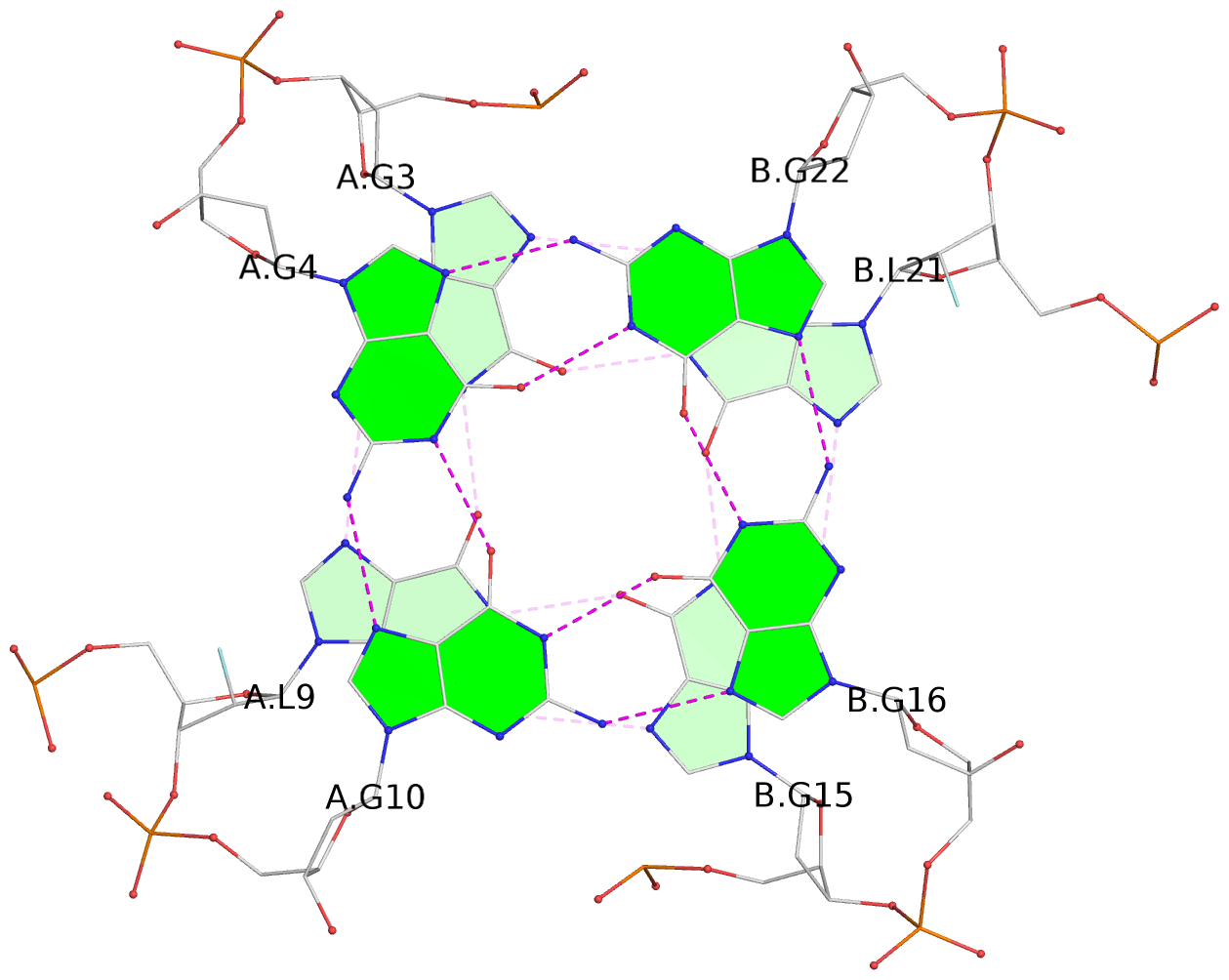
List of 3 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.336 type=other nts=4 GgGg A.DG3,A.GFL9,B.DG15,B.GFL21 2 glyco-bond=---- sugar=---- groove=---- planarity=0.143 type=planar nts=4 GGGG A.DG4,A.DG10,B.DG16,B.DG22 3 glyco-bond=---- sugar=--.- groove=---- planarity=0.367 type=bowl nts=4 GGGG A.DG5,A.DG11,B.DG17,B.DG23
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, inter-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.