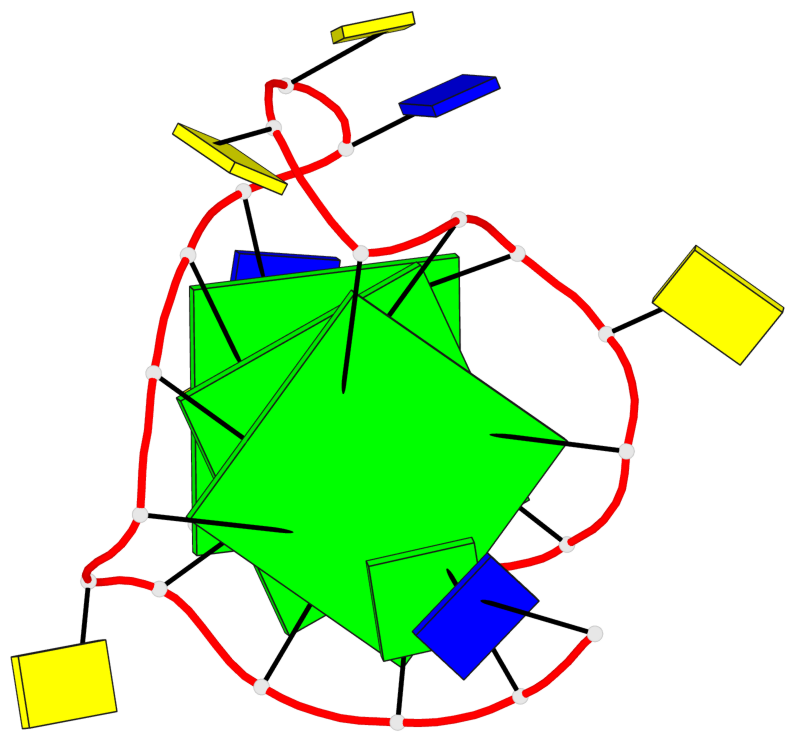
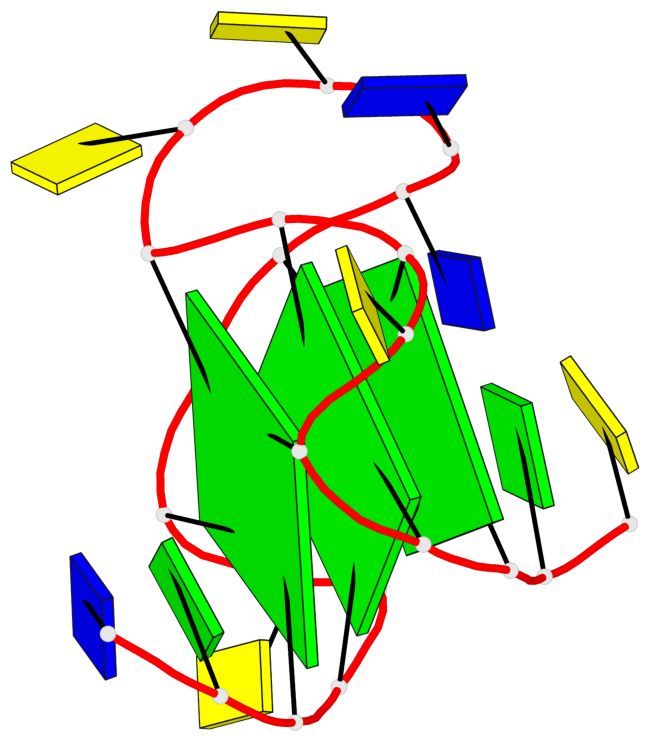
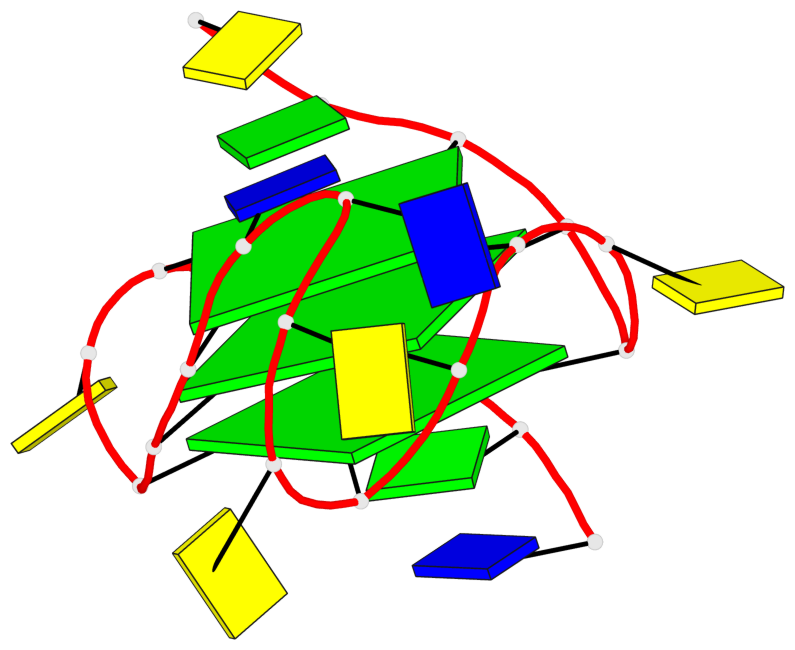
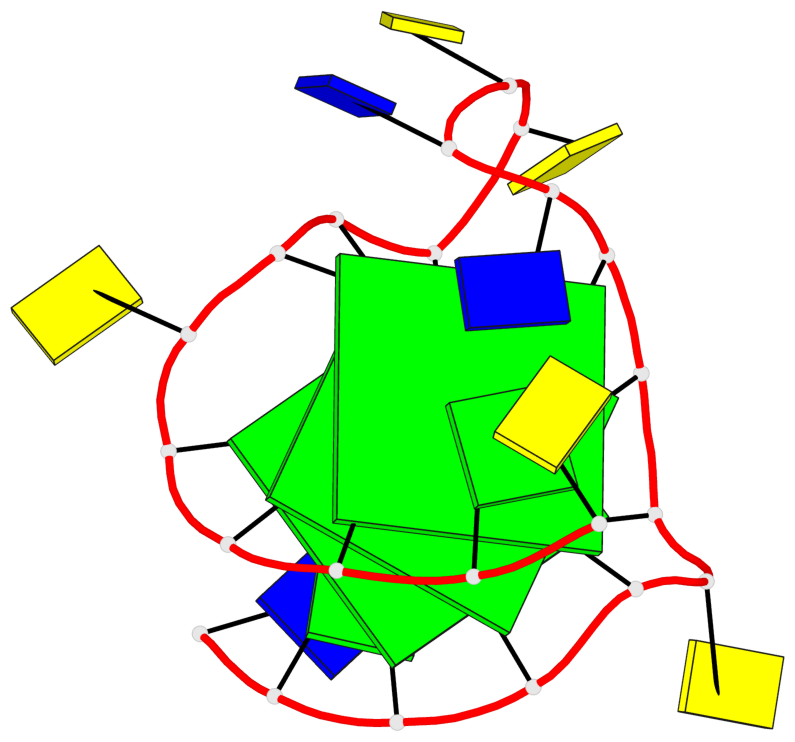
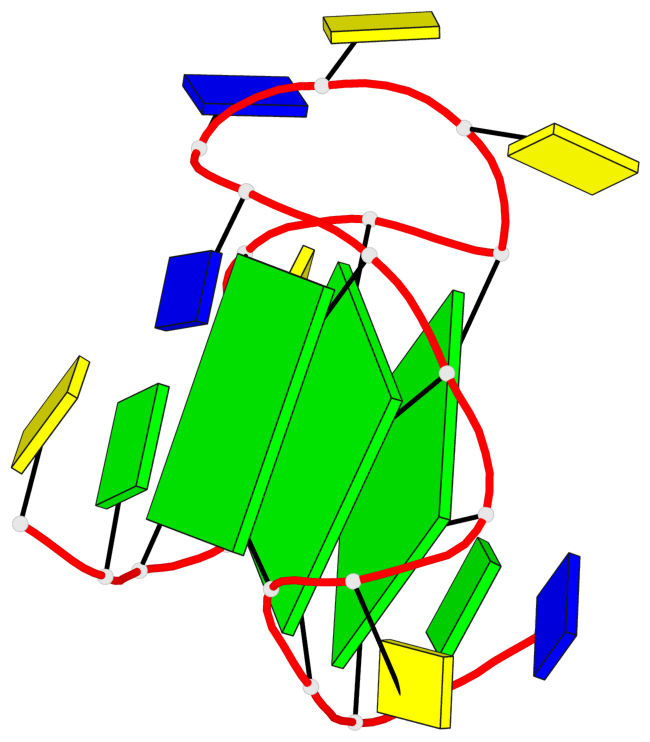
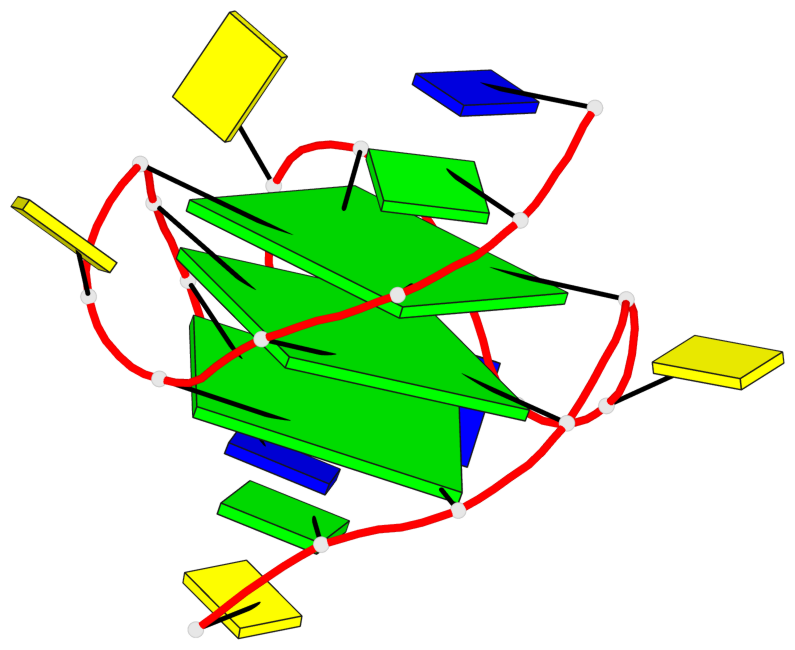
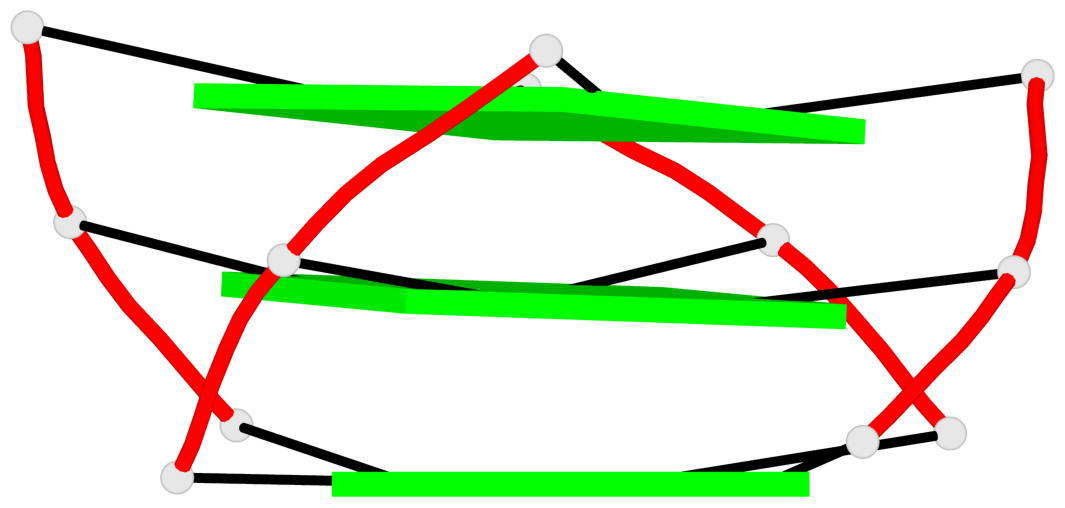
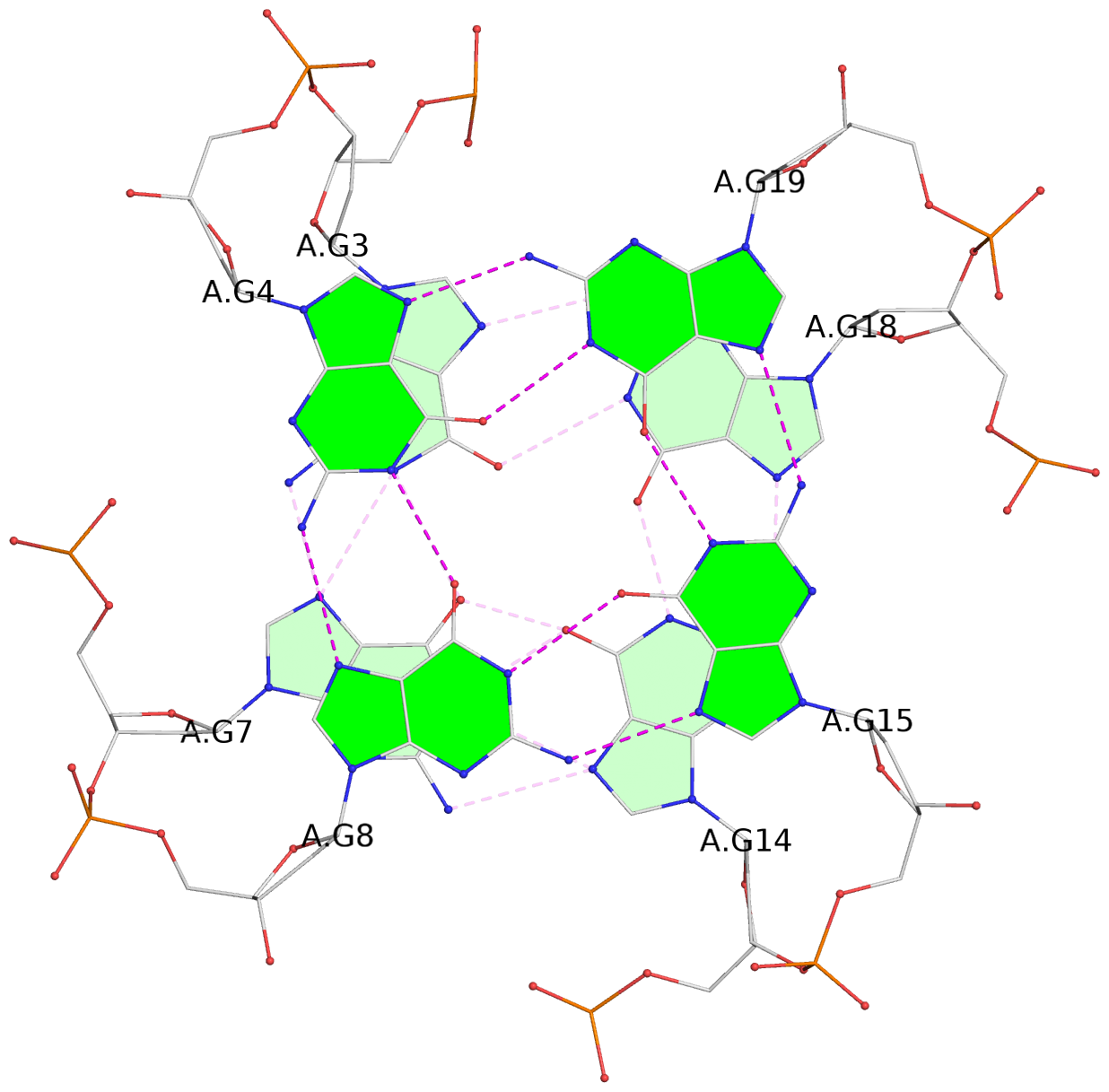
Detailed DSSR results for the G-quadruplex: PDB entry 2m27
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 2m27
- Class
- DNA
- Method
- NMR
- Summary
- Major g-quadruplex structure formed in human vegf promoter, a monomeric parallel-stranded quadruplex
- Reference
- Agrawal P, Hatzakis E, Guo K, Carver M, Yang D (2013): "Solution structure of the major G-quadruplex formed in the human VEGF promoter in K+: insights into loop interactions of the parallel G-quadruplexes." Nucleic Acids Res., 41, 10584-10592. doi: 10.1093/nar/gkt784.
- Abstract
- G4 notes
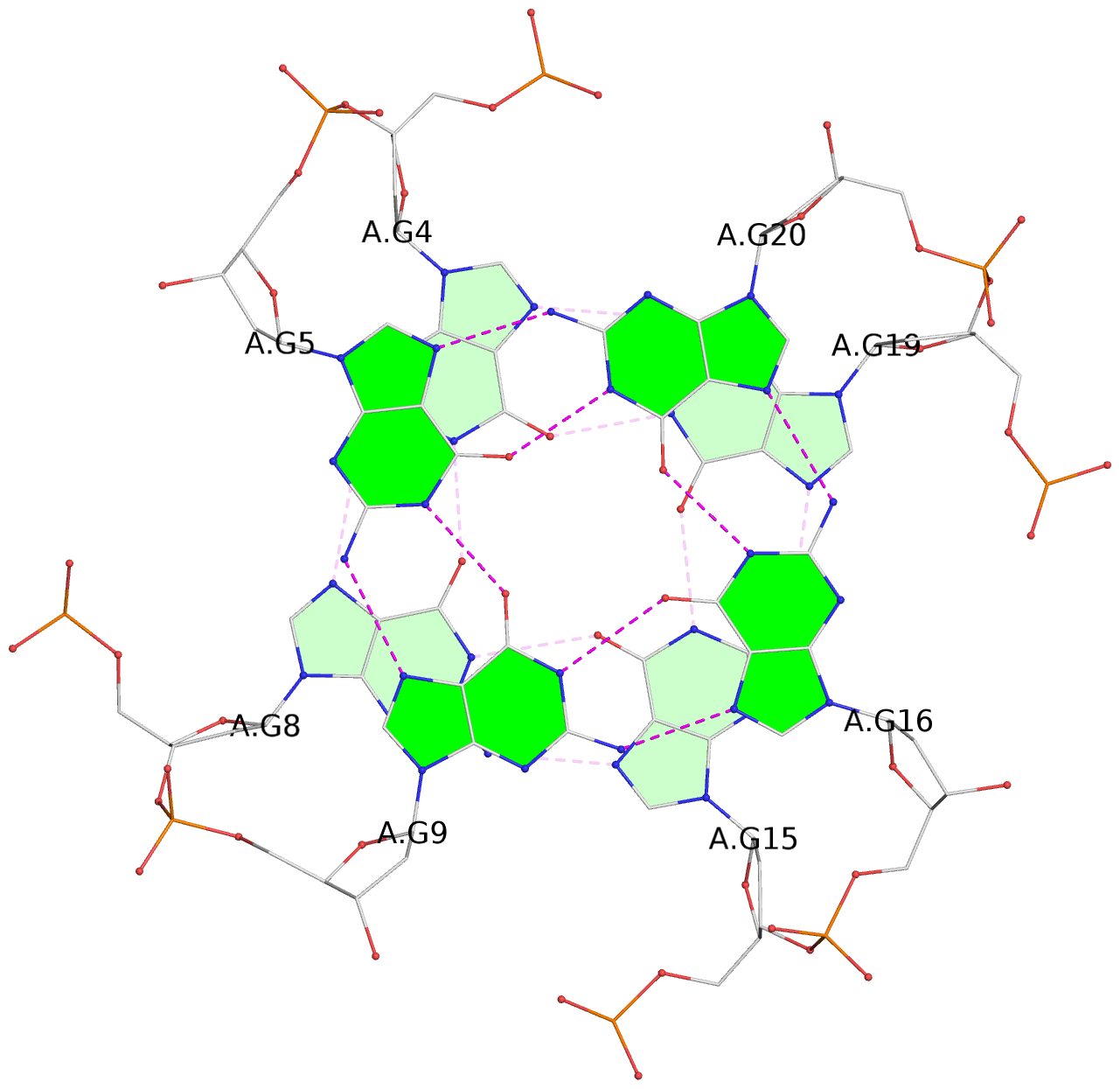
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(-P-P-P), parallel(4+0), UUUU
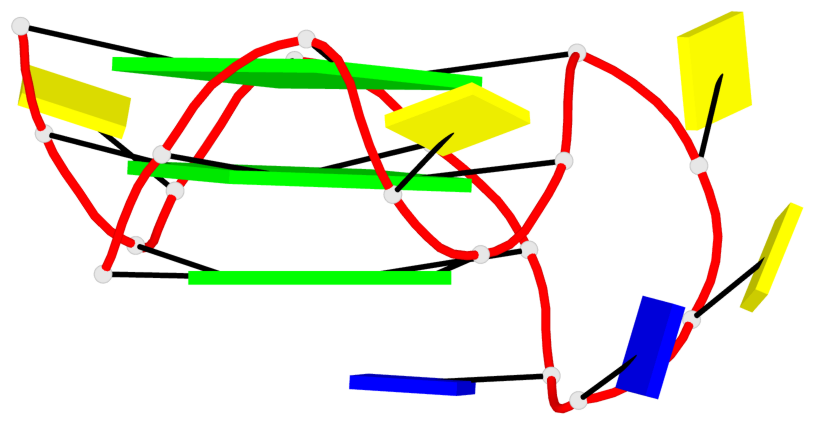
Base-block schematics in six views
List of 3 G-tetrads
1 glyco-bond=---- sugar=.... groove=---- planarity=0.062 type=planar nts=4 GGGG A.DG3,A.DG7,A.DG14,A.DG18 2 glyco-bond=---- sugar=.... groove=---- planarity=0.032 type=planar nts=4 GGGG A.DG4,A.DG8,A.DG15,A.DG19 3 glyco-bond=---- sugar=.... groove=---- planarity=0.089 type=planar nts=4 GGGG A.DG5,A.DG9,A.DG16,A.DG20
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.