Detailed DSSR results for the G-quadruplex: PDB entry 2m4p
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 2m4p
- Class
- DNA
- Method
- NMR
- Summary
- Solution structure of an intramolecular propeller-type g-quadruplex containing a single bulge
- Reference
- Mukundan VT, Phan AT (2013): "Bulges in G-Quadruplexes: Broadening the Definition of G-Quadruplex-Forming Sequences." J.Am.Chem.Soc., 135, 5017-5028. doi: 10.1021/ja310251r.
- Abstract
- We report on the first solution structure of an intramolecular G-quadruplex containing a single bulge and present evidence for extensive occurrence of bulges in different G-quadruplex contexts. The NMR solution structure of the d(TTGTGGTGGGTGGGTGGGT) sequence reveals a propeller-type parallel-stranded G-quadruplex containing three G-tetrad layers, three double-chain-reversal loops, and a bulge. All guanines participate in the formation of the G-tetrad core, despite the interruption between the first guanine and the next two guanines by a thymine, which forms a single-residue bulge and is projected out of the G-tetrad core. To provide a more general understanding about the formation of bulges within G-quadruplexes, we systematically investigated the effects of the residue type, the size, the position, and the number of bulges on the structure and stability of G-quadruplexes. The formation of bulges has also been observed in two different G-quadruplex scaffolds with different strand orientations and folding topologies. Our results show that bulges can be formed in many different situations within G-quadruplexes. While many sequences tested in this study can form stable G-quadruplex structures, all of them defy the description of sequences G3+NL1G3+NL2G3+NL3G3+, currently used in most bioinformatics searches for identifying potential G-quadruplex-forming sequences in the genomes. Broadening of this description to include the possibilities of bulge formation should allow the identification of more G-quadruplex-forming sequences which went unnoticed in the earlier searches. This study could also open the possibilities of exploiting bulges as recognition elements for interactions between G-quadruplexes and other molecules.
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(-P-P-P), parallel(4+0), UUUU
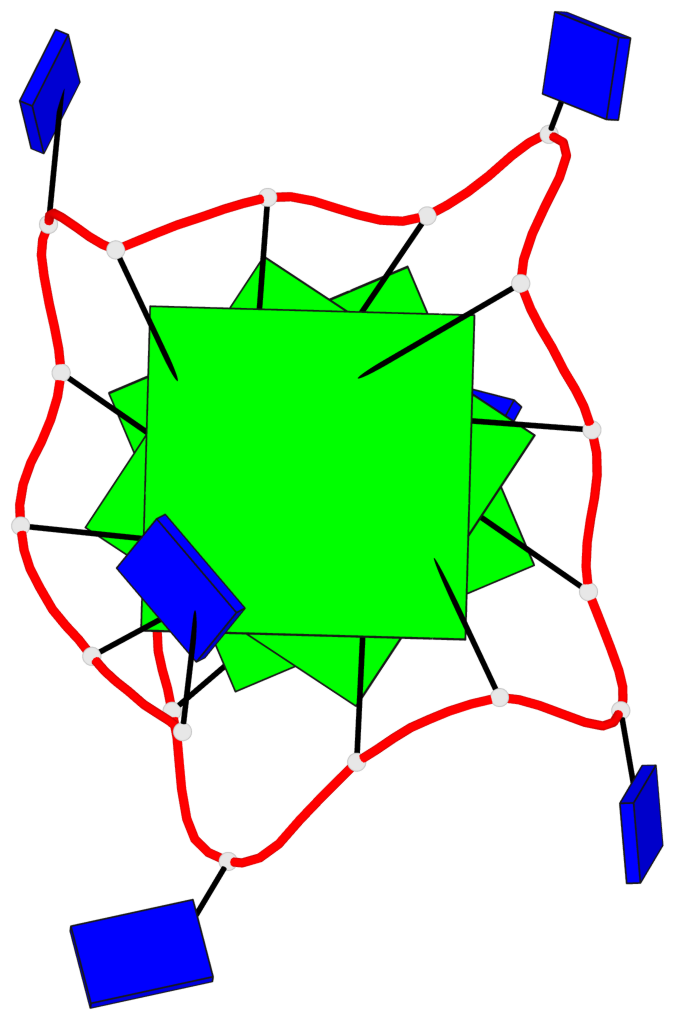
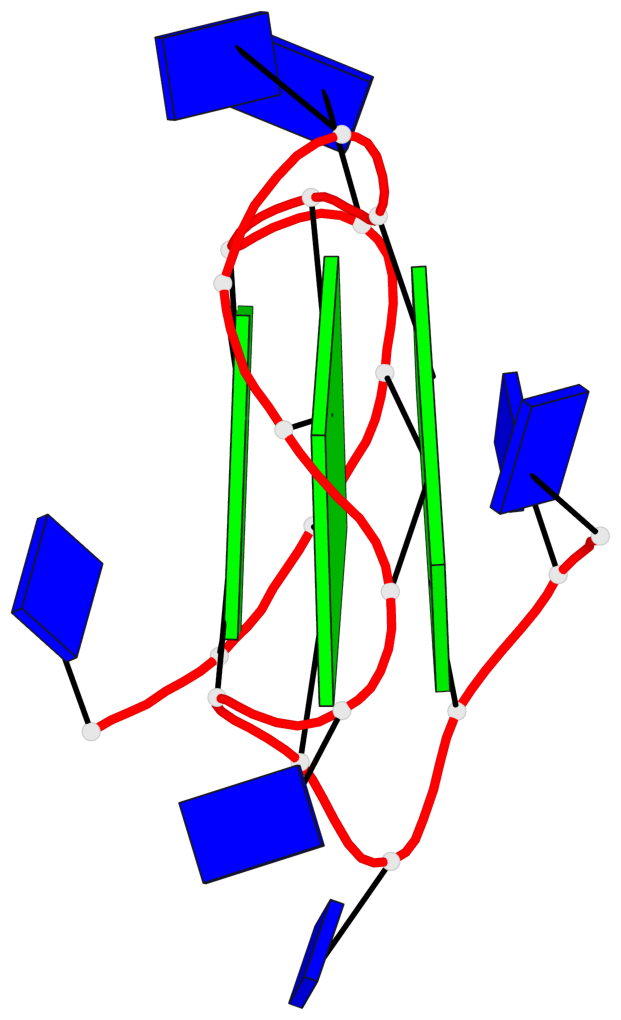
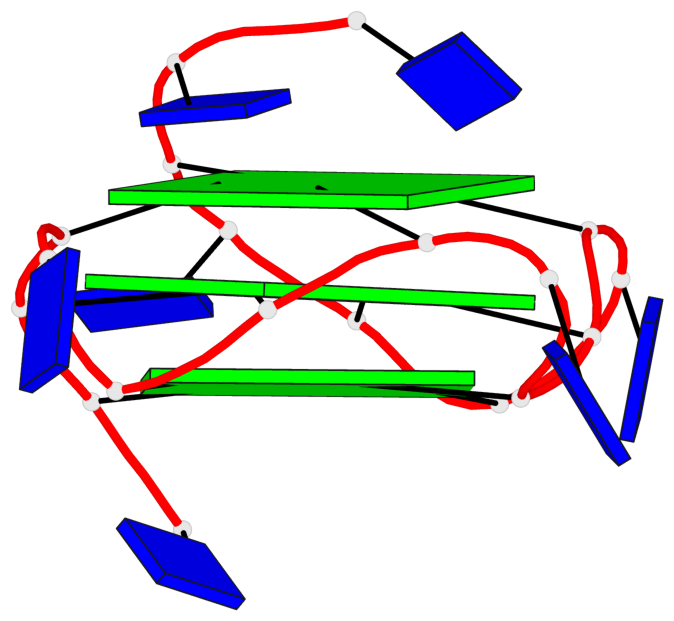
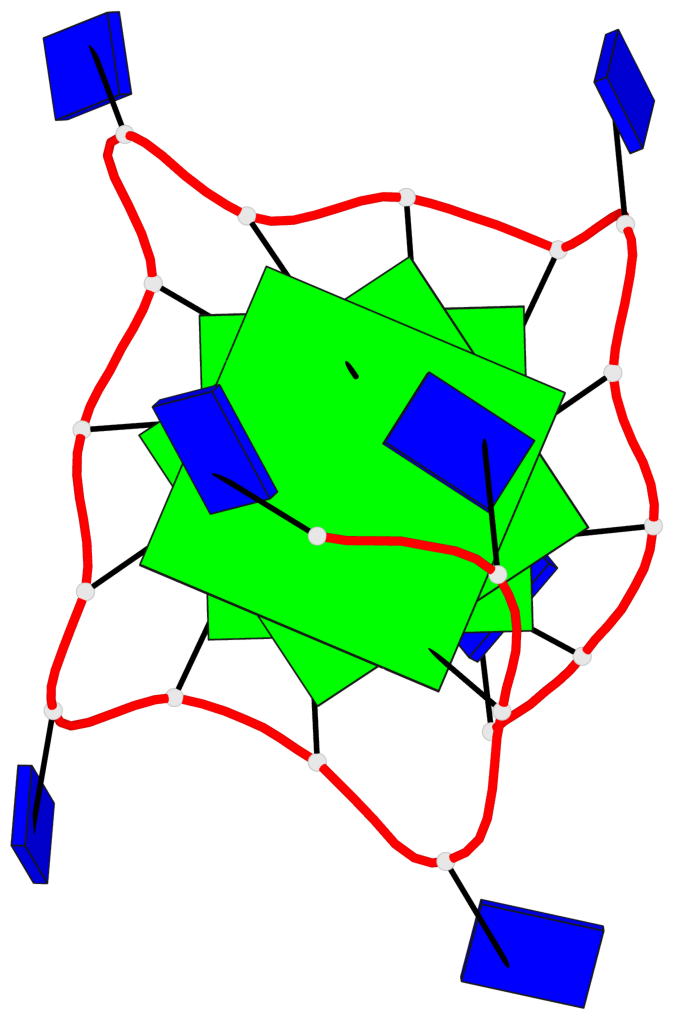
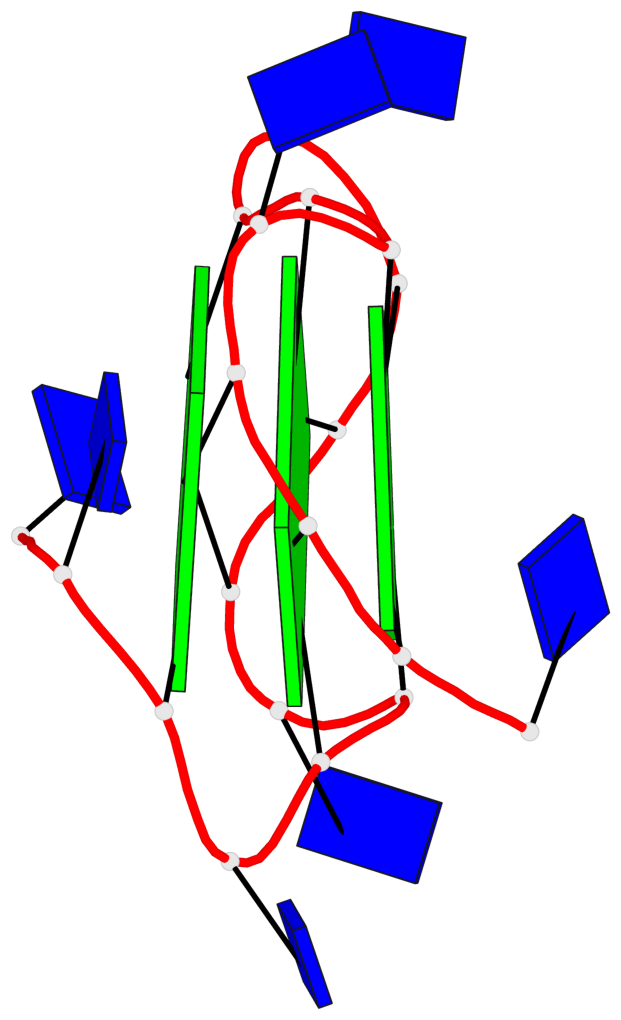
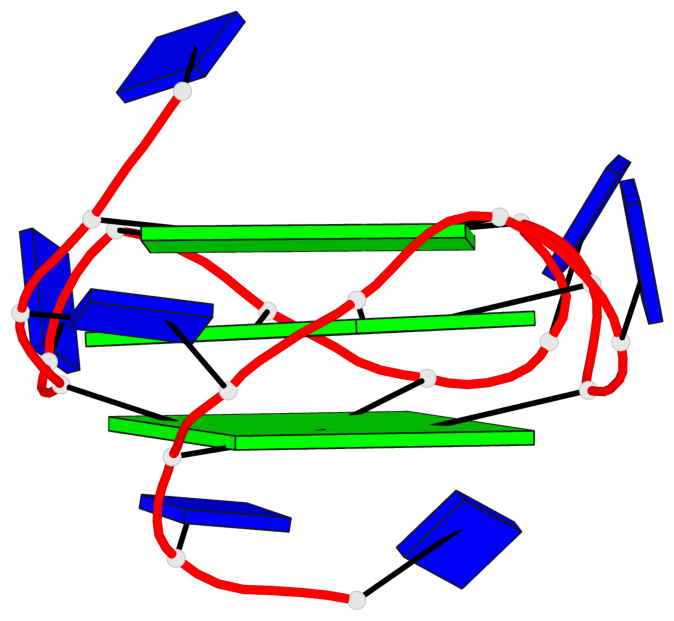
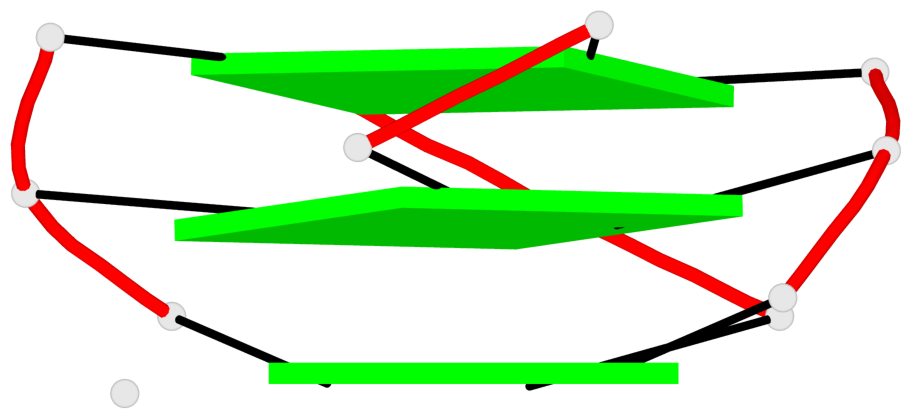
Base-block schematics in six views
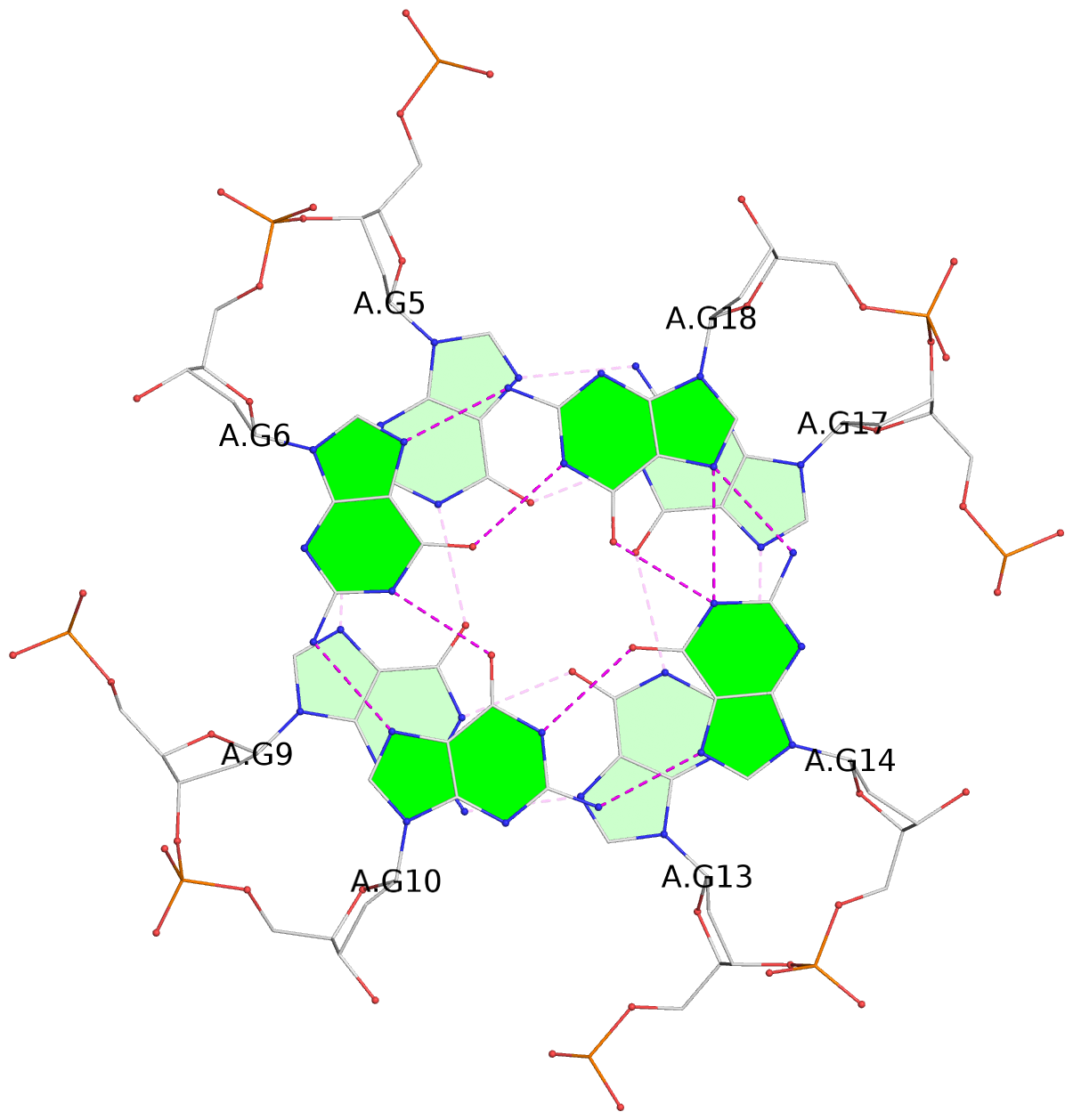
List of 3 G-tetrads
1 glyco-bond=---- sugar=..-. groove=---- planarity=0.145 type=planar nts=4 GGGG A.DG3,A.DG8,A.DG12,A.DG16 2 glyco-bond=---- sugar=---- groove=---- planarity=0.084 type=planar nts=4 GGGG A.DG5,A.DG9,A.DG13,A.DG17 3 glyco-bond=---- sugar=---- groove=---- planarity=0.087 type=planar nts=4 GGGG A.DG6,A.DG10,A.DG14,A.DG18
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.